Metagenomic profiling of a microbial assemblage associated with the California mussel: a node in networks of carbon and nitrogen cycling
- PMID: 20463896
- PMCID: PMC2865538
- DOI: 10.1371/journal.pone.0010518
Metagenomic profiling of a microbial assemblage associated with the California mussel: a node in networks of carbon and nitrogen cycling
Abstract
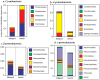
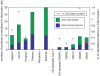
Mussels are conspicuous and often abundant members of rocky shores and may constitute an important site for the nitrogen cycle due to their feeding and excretion activities. We used shotgun metagenomics of the microbial community associated with the surface of mussels (Mytilus californianus) on Tatoosh Island in Washington state to test whether there is a nitrogen-based microbial assemblage associated with mussels. Analyses of both tidepool mussels and those on emergent benches revealed a diverse community of Bacteria and Archaea with approximately 31 million bp from 6 mussels in each habitat. Using MG-RAST, between 22.5-25.6% were identifiable using the SEED non-redundant database for proteins. Of those fragments that were identifiable through MG-RAST, the composition was dominated by Cyanobacteria and Alpha- and Gamma-proteobacteria. Microbial composition was highly similar between the tidepool and emergent bench mussels, suggesting similar functions across these different microhabitats. One percent of the proteins identified in each sample were related to nitrogen cycling. When normalized to protein discovery rate, the high diversity and abundance of enzymes related to the nitrogen cycle in mussel-associated microbes is as great or greater than that described for other marine metagenomes. In some instances, the nitrogen-utilizing profile of this assemblage was more concordant with soil metagenomes in the Midwestern U.S. than for open ocean system. Carbon fixation and Calvin cycle enzymes further represented 0.65 and 1.26% of all proteins and their abundance was comparable to a number of open ocean marine metagenomes. In sum, the diversity and abundance of nitrogen and carbon cycle related enzymes in the microbes occupying the shells of Mytilus californianus suggest these mussels provide a node for microbial populations and thus biogeochemical processes.
Conflict of interest statement
Figures


References
-
- Dugdale RC, Goering JJ. Uptake of new and regenerated forms of nitrogen in primary productivity. Limnol Oceanogr. 1967;12:196–206.
-
- Eppley RW, Reuger EH, Venrick EL, Mullin M. A study of plankton dynamics and nutrient cycling in the Central Gyre of the North Pacific Ocean. Limnol Oceanogr. 1973;18:534–551.
-
- Bode A, Barquero S, Gonzalez N, Alvarez-Ossorio M, Varela M. Contribution of heterotrophic plankton to nitrogen regeneration in the upwelling ecosystem of A. Coruña (NW Spain). J Plankton Res. 2004;26:11–28.
-
- Wootton JT. Direct and indirect effects of nutrients on intertidal community structure: variable consequences of seabird guano. J Expl Mar Biol Ecol. 1991;151:139–153.
-
- Bracken MES. Invertebrate-mediated nutrient loading increases growth of an intertidal macroalga. J Phycol. 2004;40:1032–1041.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

