Transcriptional and translational regulatory responses to iron limitation in the globally distributed marine bacterium Candidatus pelagibacter ubique
- PMID: 20463970
- PMCID: PMC2864753
- DOI: 10.1371/journal.pone.0010487
Transcriptional and translational regulatory responses to iron limitation in the globally distributed marine bacterium Candidatus pelagibacter ubique
Abstract
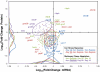
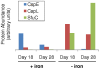
Iron is recognized as an important micronutrient that limits microbial plankton productivity over vast regions of the oceans. We investigated the gene expression responses of Candidatus Pelagibacter ubique cultures to iron limitation in natural seawater media supplemented with a siderophore to chelate iron. Microarray data indicated transcription of the periplasmic iron binding protein sfuC increased by 16-fold, and iron transporter subunits, iron-sulfur center assembly genes, and the putative ferroxidase rubrerythrin transcripts increased to a lesser extent. Quantitative peptide mass spectrometry revealed that sfuC protein abundance increased 27-fold, despite an average decrease of 59% across the global proteome. Thus, we propose sfuC as a marker gene for indicating iron limitation in marine metatranscriptomic and metaproteomic ecological surveys. The marked proteome reduction was not directly correlated to changes in the transcriptome, implicating post-transcriptional regulatory mechanisms as modulators of protein expression. Two RNA-binding proteins, CspE and CspL, correlated well with iron availability, suggesting that they may contribute to the observed differences between the transcriptome and proteome. We propose a model in which the RNA-binding activity of CspE and CspL selectively enables protein synthesis of the iron acquisition protein SfuC during transient growth-limiting episodes of iron scarcity.
Conflict of interest statement
Figures


References
-
- Martin JH, Gordon RM, Fitzwater SE. Iron in Antarctic waters. Nature. 1990;345:156–158. doi: 10.1038/345156a0. - DOI
-
- Coale KH, Johnson KS, Fitzwater SE, Gordon RM, Tanner S, et al. A massive phytoplankton bloom induced by an ecosystem-scale iron fertilization experiment in the equatorial Pacific Ocean. Nature. 1996;383:495–501. - PubMed
-
- Boyd PW, Watson AJ, Law CS, Abraham ER, Trull T, et al. A mesoscale phytoplankton bloom in the polar Southern Ocean stimulated by iron fertilization. Nature. 2000;407:695–702. - PubMed
-
- Fung IY, Meyn SK, Tegen I, Doney SC, John JG, et al. Iron supply and demand in the upper ocean. Global Biogeochem Cycles. 2000;14:281–295.
-
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

