Molecular analysis of endothelial progenitor cell (EPC) subtypes reveals two distinct cell populations with different identities
- PMID: 20465783
- PMCID: PMC2881111
- DOI: 10.1186/1755-8794-3-18
Molecular analysis of endothelial progenitor cell (EPC) subtypes reveals two distinct cell populations with different identities
Abstract
Background: The term endothelial progenitor cells (EPCs) is currently used to refer to cell populations which are quite dissimilar in terms of biological properties. This study provides a detailed molecular fingerprint for two EPC subtypes: early EPCs (eEPCs) and outgrowth endothelial cells (OECs).
Methods: Human blood-derived eEPCs and OECs were characterised by using genome-wide transcriptional profiling, 2D protein electrophoresis, and electron microscopy. Comparative analysis at the transcript and protein level included monocytes and mature endothelial cells as reference cell types.
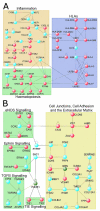
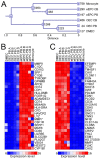
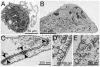
Results: Our data show that eEPCs and OECs have strikingly different gene expression signatures. Many highly expressed transcripts in eEPCs are haematopoietic specific (RUNX1, WAS, LYN) with links to immunity and inflammation (TLRs, CD14, HLAs), whereas many transcripts involved in vascular development and angiogenesis-related signalling pathways (Tie2, eNOS, Ephrins) are highly expressed in OECs. Comparative analysis with monocytes and mature endothelial cells clusters eEPCs with monocytes, while OECs segment with endothelial cells. Similarly, proteomic analysis revealed that 90% of spots identified by 2-D gel analysis are common between OECs and endothelial cells while eEPCs share 77% with monocytes. In line with the expression pattern of caveolins and cadherins identified by microarray analysis, ultrastructural evaluation highlighted the presence of caveolae and adherens junctions only in OECs.
Conclusions: This study provides evidence that eEPCs are haematopoietic cells with a molecular phenotype linked to monocytes; whereas OECs exhibit commitment to the endothelial lineage. These findings indicate that OECs might be an attractive cell candidate for inducing therapeutic angiogenesis, while eEPC should be used with caution because of their monocytic nature.
Figures



References
-
- Tateishi-Yuyama E, Matsubara H, Murohara T, Ikeda U, Shintani S, Masaki H, Amano K, Kishimoto Y, Yoshimoto K, Akashi H. Therapeutic angiogenesis for patients with limb ischaemia by autologous transplantation of bone-marrow cells: a pilot study and a randomised controlled trial. Lancet. 2002;360(9331):427–435. doi: 10.1016/S0140-6736(02)09670-8. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

