Toward an integrated structural model of the 26S proteasome
- PMID: 20467039
- PMCID: PMC2938054
- DOI: 10.1074/mcp.R000002-MCP201
Toward an integrated structural model of the 26S proteasome
Abstract
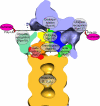
The 26S proteasome is the end point of the ubiquitin-proteasome pathway and degrades ubiquitylated substrates. It is composed of the 20S core particle (CP), where degradation occurs, and the 19S regulatory particle (RP), which ensures substrate specificity of degradation. Whereas the CP is resolved to atomic resolution, the architecture of the RP is largely unknown. We provide a comprehensive analysis of the current structural knowledge on the RP, including structures of the RP subunits, physical protein-protein interactions, and cryoelectron microscopy data. These data allowed us to compute an atomic model for the CP-AAA-ATPase subcomplex. In addition to this atomic model, further subunits can be mapped approximately, which lets us hypothesize on the substrate path during its degradation.
Figures


Similar articles
-
Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach.Proc Natl Acad Sci U S A. 2012 Jan 31;109(5):1380-7. doi: 10.1073/pnas.1120559109. Epub 2012 Jan 23. Proc Natl Acad Sci U S A. 2012. PMID: 22307589 Free PMC article.
-
An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome.Biochem Biophys Res Commun. 2009 Oct 16;388(2):228-33. doi: 10.1016/j.bbrc.2009.07.145. Epub 2009 Aug 3. Biochem Biophys Res Commun. 2009. PMID: 19653995 Free PMC article.
-
Structural insights into the functional cycle of the ATPase module of the 26S proteasome.Proc Natl Acad Sci U S A. 2017 Feb 7;114(6):1305-1310. doi: 10.1073/pnas.1621129114. Epub 2017 Jan 23. Proc Natl Acad Sci U S A. 2017. PMID: 28115689 Free PMC article.
-
Proteasome in action: substrate degradation by the 26S proteasome.Biochem Soc Trans. 2021 Apr 30;49(2):629-644. doi: 10.1042/BST20200382. Biochem Soc Trans. 2021. PMID: 33729481 Free PMC article. Review.
-
Molecular architecture and assembly of the eukaryotic proteasome.Annu Rev Biochem. 2013;82:415-45. doi: 10.1146/annurev-biochem-060410-150257. Epub 2013 Mar 13. Annu Rev Biochem. 2013. PMID: 23495936 Free PMC article. Review.
Cited by
-
Aggrephagy: selective disposal of protein aggregates by macroautophagy.Int J Cell Biol. 2012;2012:736905. doi: 10.1155/2012/736905. Epub 2012 Mar 22. Int J Cell Biol. 2012. PMID: 22518139 Free PMC article.
-
Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach.Proc Natl Acad Sci U S A. 2012 Jan 31;109(5):1380-7. doi: 10.1073/pnas.1120559109. Epub 2012 Jan 23. Proc Natl Acad Sci U S A. 2012. PMID: 22307589 Free PMC article.
-
Go hybrid: EM, crystallography, and beyond.Curr Opin Struct Biol. 2012 Oct;22(5):627-35. doi: 10.1016/j.sbi.2012.07.006. Epub 2012 Jul 24. Curr Opin Struct Biol. 2012. PMID: 22835744 Free PMC article. Review.
-
Ubiquitin ligases of the N-end rule pathway: assessment of mutations in UBR1 that cause the Johanson-Blizzard syndrome.PLoS One. 2011;6(9):e24925. doi: 10.1371/journal.pone.0024925. Epub 2011 Sep 13. PLoS One. 2011. PMID: 21931868 Free PMC article.
-
The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together.Proc Natl Acad Sci U S A. 2012 Jan 3;109(1):149-54. doi: 10.1073/pnas.1117648108. Epub 2011 Dec 20. Proc Natl Acad Sci U S A. 2012. PMID: 22187461 Free PMC article.
References
-
- Murata S., Yashiroda H., Tanaka K. (2009) Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115 - PubMed
-
- Glickman M. H., Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 - PubMed
-
- Voges D., Zwickl P., Baumeister W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068 - PubMed
-
- Jentsch S., Haendler B. (2009) The Ubiquitin System in Health and Disease, Springer, Heidelberg, Germany - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

