Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals changes in the cytoplasmic, nuclear, and nucleolar proteomes in Vero cells infected with the coronavirus infectious bronchitis virus
- PMID: 20467043
- PMCID: PMC2938107
- DOI: 10.1074/mcp.M900345-MCP200
Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals changes in the cytoplasmic, nuclear, and nucleolar proteomes in Vero cells infected with the coronavirus infectious bronchitis virus
Abstract
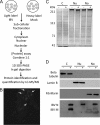
Virus-host interactions involve complex interplay between viral and host factors, rendering them an ideal target for proteomic analysis. Here we detail a high throughput quantitative proteomics analysis of Vero cells infected with the coronavirus infectious bronchitis virus (IBV), a positive strand RNA virus that replicates in the cytoplasm. Stable isotope labeling with amino acids in cell culture (SILAC) was used in conjunction with LC-MS/MS to identify and quantify 1830 cellular and two viral proteins from IBV-infected cells. Fractionation of cells into cytoplasmic, nuclear, and nucleolar extracts was used to reduce sample complexity and provide information on the trafficking of proteins between the different compartments. Each fraction showed a proportion of proteins exhibiting >or=2-fold changes in abundance. Ingenuity Pathway Analysis revealed that proteins that changed in response to infection could be grouped into different functional categories. These included proteins regulated by NF-kappaB- and AP-1-dependent pathways and proteins involved in the cytoskeleton and molecular motors. A luciferase-based reporter gene assay was used to validate the up-regulation of AP-1- and NF-kappaB-dependent transcription in IBV-infected cells and confirmed using immunofluorescence. Immunofluorescence was used to validate changes in the subcellular localization of vimentin and myosin VI in IBV-infected cells. The proteomics analysis also confirmed the presence of the viral nucleocapsid protein as localizing in the cytoplasm, nucleus, and nucleolus and the viral membrane protein in the cytoplasmic fraction. This research is the first application of SILAC to study total host cell proteome changes in response to positive sense RNA virus infection and illustrates the versatility of this technique as applied to infectious disease research.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

