Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage
- PMID: 20471290
- PMCID: PMC3129685
- DOI: 10.1016/j.immuni.2010.04.016
Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage
Abstract
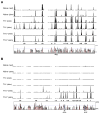
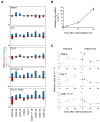
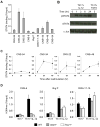
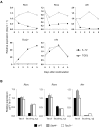
Phenotypic plasticity of T helper 17 (Th17) cells suggests instability of chromatin structure of key genes of this lineage. We identified epigenetic modifications across the clustered Il17a and Il17f and the Ifng loci before and after differential IL-12 or TGF-beta cytokine signaling, which induce divergent fates of Th17 cell precursors. We found that Th17 cell precursors had substantial remodeling of the Ifng locus, but underwent critical additional modifications to enable high expression when stimulated by IL-12. Permissive modifications across the Il17a-Il17f locus were amplified by TGF-beta signaling in Th17 cells, but were rapidly reversed downstream of IL-12-induced silencing of the Rorc gene by the transcription factors STAT4 and T-bet. These findings reveal substantial chromatin instability of key transcription factor and cytokine genes of Th17 cells and support a model of Th17 cell lineage plasticity in which cell-extrinsic factors modulate Th17 cell fates through differential effects on the epigenetic status of Th17 cell lineage factors.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Keeping one's option open.Immunity. 2010 May 28;32(5):581-3. doi: 10.1016/j.immuni.2010.05.008. Immunity. 2010. PMID: 20510865
References
-
- Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. - PubMed
-
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. - PubMed
-
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

