Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination
- PMID: 20471950
- PMCID: PMC3132514
- DOI: 10.1016/j.molcel.2010.02.032
Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination
Abstract
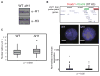
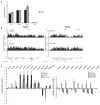
How polycomb group proteins repress gene expression in vivo is not known. While histone-modifying activities of the polycomb repressive complexes (PRCs) have been studied extensively, in vitro data have suggested a direct activity of the PRC1 complex in compacting chromatin. Here, we investigate higher-order chromatin compaction of polycomb targets in vivo. We show that PRCs are required to maintain a compact chromatin state at Hox loci in embryonic stem cells (ESCs). There is specific decompaction in the absence of PRC2 or PRC1. This is due to a PRC1-like complex, since decompaction occurs in Ring1B null cells that still have PRC2-mediated H3K27 methylation. Moreover, we show that the ability of Ring1B to restore a compact chromatin state and to repress Hox gene expression is not dependent on its histone ubiquitination activity. We suggest that Ring1B-mediated chromatin compaction acts to directly limit transcription in vivo.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Chromatin compaction at Hox loci: a polycomb tale beyond histone tails.Mol Cell. 2010 May 14;38(3):321-2. doi: 10.1016/j.molcel.2010.04.018. Mol Cell. 2010. PMID: 20471938
References
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. - PubMed
-
- Bates DL, Butler PJ, Pearson EC, Thomas JO. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur J Biochem. 1981;119:469–476. - PubMed
-
- Bel-Vialar S, Itasaki N, Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129:5103–5115. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

