The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell
- PMID: 20472542
- PMCID: PMC2887050
- DOI: 10.1093/bioinformatics/btq243
The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell
Abstract
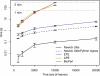
Summary: We present a suite of Unix shell programs for processing any number of phylogenetic trees of any size. They perform frequently-used tree operations without requiring user interaction. They also allow tree drawing as scalable vector graphics (SVG), suitable for high-quality presentations and further editing, and as ASCII graphics for command-line inspection. As an example we include an implementation of bootscanning, a procedure for finding recombination breakpoints in viral genomes.
Availability: C source code, Python bindings and executables for various platforms are available from http://cegg.unige.ch/newick_utils. The distribution includes a manual and example data. The package is distributed under the BSD License.
Contact: thomas.junier@unige.ch
Figures
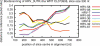
References
-
- Archie J, et al. 1986. http://evolution.genetics.washington.edu/phylip/newicktree.html.
-
- Felsenstein J. PHYLIP - Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166.
-
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

