The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation
- PMID: 20473272
- PMCID: PMC2892366
- DOI: 10.1038/emboj.2010.81
The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation
Abstract
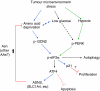
The transcription factor ATF4 regulates the expression of genes involved in amino acid metabolism, redox homeostasis and ER stress responses, and it is overexpressed in human solid tumours, suggesting that it has an important function in tumour progression. Here, we report that inhibition of ATF4 expression blocked proliferation and survival of transformed cells, despite an initial activation of cytoprotective macroautophagy. Knockdown of ATF4 significantly reduced the levels of asparagine synthetase (ASNS) and overexpression of ASNS or supplementation of asparagine in trans, reversed the proliferation block and increased survival in ATF4 knockdown cells. Both amino acid and glucose deprivation, stresses found in solid tumours, activated the upstream eukaryotic initiation factor 2alpha (eIF2alpha) kinase GCN2 to upregulate ATF4 target genes involved in amino acid synthesis and transport. GCN2 activation/overexpression and increased phospho-eIF2alpha were observed in human and mouse tumours compared with normal tissues and abrogation of ATF4 or GCN2 expression significantly inhibited tumour growth in vivo. We conclude that the GCN2-eIF2alpha-ATF4 pathway is critical for maintaining metabolic homeostasis in tumour cells, making it a novel and attractive target for anti-tumour approaches.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Comment in
-
How do tumours adapt to nutrient stress?EMBO J. 2010 Jun 16;29(12):1946-7. doi: 10.1038/emboj.2010.110. EMBO J. 2010. PMID: 20551969 Free PMC article. No abstract available.
References
-
- Ameri K, Lewis CE, Raida M, Sowter H, Hai T, Harris AL (2004) Anoxic induction of ATF-4 through HIF-1-independent pathways of protein stabilization in human cancer cells. Blood 103: 1876–1882 - PubMed
-
- Anthony TG, McDaniel BJ, Byerley RL, McGrath BC, Cavener DR, McNurlan MA, Wek RC (2004) Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J Biol Chem 279: 36553–36561 - PubMed
-
- Cooney DA, Handschumacher RE (1970) L-asparaginase and L-asparagine metabolism. Annu Rev Pharmacol 10: 421–440 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

