Over-expression of the IGI1 leading to altered shoot-branching development related to MAX pathway in Arabidopsis
- PMID: 20473553
- PMCID: PMC2898107
- DOI: 10.1007/s11103-010-9645-0
Over-expression of the IGI1 leading to altered shoot-branching development related to MAX pathway in Arabidopsis
Abstract
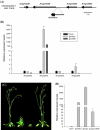
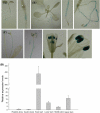
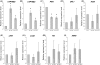
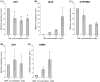
Shoot branching and growth are controlled by phytohormones such as auxin and other components in Arabidopsis. We identified a mutant (igi1) showing decreased height and bunchy branching patterns. The phenotypes reverted to the wild type in response to RNA interference with the IGI1 gene. Histochemical analysis by GUS assay revealed tissue-specific gene expression in the anther and showed that the expression levels of the IGI1 gene in apical parts, including flowers, were higher than in other parts of the plants. The auxin biosynthesis component gene, CYP79B2, was up-regulated in igi1 mutants and the IGI1 gene was down-regulated by IAA treatment. These results indicated that there is an interplay regulation between IGI1 and phytohormone auxin. Moreover, the expression of the auxin-related shoot branching regulation genes, MAX3 and MAX4, was down-regulated in igi1 mutants. Taken together, these results indicate that the overexpression of the IGI1 influenced MAX pathway in the shoot branching regulation.
Figures


References
-
- Bangerth F. Response of cytokinin concentration in the xylem exudate of bean (Phaseolus vulgaris L.) plants to decapitation and auxin treatment, and relationship to apical dominance. Planta. 1994;194:439–442. doi: 10.1007/BF00197546. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

