Chem2Bio2RDF: a semantic framework for linking and data mining chemogenomic and systems chemical biology data
- PMID: 20478034
- PMCID: PMC2881087
- DOI: 10.1186/1471-2105-11-255
Chem2Bio2RDF: a semantic framework for linking and data mining chemogenomic and systems chemical biology data
Abstract
Background: Recently there has been an explosion of new data sources about genes, proteins, genetic variations, chemical compounds, diseases and drugs. Integration of these data sources and the identification of patterns that go across them is of critical interest. Initiatives such as Bio2RDF and LODD have tackled the problem of linking biological data and drug data respectively using RDF. Thus far, the inclusion of chemogenomic and systems chemical biology information that crosses the domains of chemistry and biology has been very limited
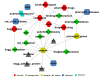
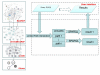
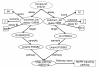
Results: We have created a single repository called Chem2Bio2RDF by aggregating data from multiple chemogenomics repositories that is cross-linked into Bio2RDF and LODD. We have also created a linked-path generation tool to facilitate SPARQL query generation, and have created extended SPARQL functions to address specific chemical/biological search needs. We demonstrate the utility of Chem2Bio2RDF in investigating polypharmacology, identification of potential multiple pathway inhibitors, and the association of pathways with adverse drug reactions.
Conclusions: We have created a new semantic systems chemical biology resource, and have demonstrated its potential usefulness in specific examples of polypharmacology, multiple pathway inhibition and adverse drug reaction--pathway mapping. We have also demonstrated the usefulness of extending SPARQL with cheminformatics and bioinformatics functionality.
Figures




References
-
- Scheiber J, Chen B, Milik M, Sukuru SC, Bender A, Mikhailov D, Whitebread S, Hamon J, Azzaoui K, Urban L, Glick M, Davies JW, Jenkins JL. Gaining insight into off-target mediated effects of drug candidates with a comprehensive systems chemical biology analysis. J Chem Inf Model. 2009;49(2):308–17. doi: 10.1021/ci800344p. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

