Interactions of SARS coronavirus nucleocapsid protein with the host cell proteasome subunit p42
- PMID: 20478047
- PMCID: PMC2894783
- DOI: 10.1186/1743-422X-7-99
Interactions of SARS coronavirus nucleocapsid protein with the host cell proteasome subunit p42
Abstract
Background: Severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spreads rapidly and has a high case-mortality rate. The nucleocapsid protein (NP) of SARS-CoV may be critical for pathogenicity. This study sought to discover the host proteins that interact with SARS-CoV NP.
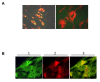
Results: Using surface plasmon resonance biomolecular interaction analysis (SPR/BIA) and matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry, we found that only the proteasome subunit p42 from human fetal lung diploid fibroblast (2BS) cells bound to SARS-CoV NP. This interaction was confirmed by the glutathione S-transferase (GST) fusion protein pulldown technique. The co-localization signal of SARS-CoV NP and proteasome subunit p42 in 2BS cells was detected using indirect immunofluorescence and confocal microscopy. p42 is a subunit of the 26S proteasome; this large, multi-protein complex is a component of the ubiquitin-proteasome pathway, which is involved in a variety of basic cellular processes and inflammatory responses.
Conclusion: To our knowledge, this is the first report that SARS-CoV NP interacts with the proteasome subunit p42 within host cells. These data enhance our understanding of the molecular mechanisms of SARS-CoV pathogenicity and the means by which SARS-CoV interacts with host cells.
Figures
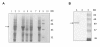
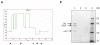
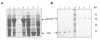

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

