ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids
- PMID: 20478830
- PMCID: PMC2896094
- DOI: 10.1093/nar/gkq399
ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids
Abstract
It is informative to detect highly conserved positions in proteins and nucleic acid sequence/structure since they are often indicative of structural and/or functional importance. ConSurf (http://consurf.tau.ac.il) and ConSeq (http://conseq.tau.ac.il) are two well-established web servers for calculating the evolutionary conservation of amino acid positions in proteins using an empirical Bayesian inference, starting from protein structure and sequence, respectively. Here, we present the new version of the ConSurf web server that combines the two independent servers, providing an easier and more intuitive step-by-step interface, while offering the user more flexibility during the process. In addition, the new version of ConSurf calculates the evolutionary rates for nucleic acid sequences. The new version is freely available at: http://consurf.tau.ac.il/.
Figures
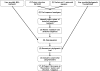

References
-
- Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. - PubMed
-
- Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. - PubMed
-
- Mayrose I, Graur D, Ben-Tal N, Pupko T. Comparison of site-specific rate-inference methods for protein sequences: empirical Bayesian methods are superior. Mol. Biol. Evol. 2004;21:1781–1791. - PubMed
-
- Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl 1):S71–S77. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

