The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells
- PMID: 20479118
- PMCID: PMC2882835
- DOI: 10.1084/jem.20100223
The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells
Abstract
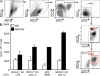
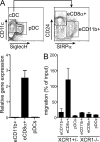
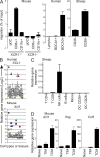
Human BDCA3+ dendritic cells (DCs) were suggested to be homologous to mouse CD8alpha+ DCs. We demonstrate that human BDCA3+ DCs are more efficient than their BDCA1+ counterparts or plasmacytoid DCs (pDCs) in cross-presenting antigen and activating CD8+ T cells, which is similar to mouse CD8alpha+ DCs as compared with CD11b+ DCs or pDCs, although with more moderate differences between human DC subsets. Yet, no specific marker was known to be shared between homologous DC subsets across species. We found that XC chemokine receptor 1 (XCR1) is specifically expressed and active in mouse CD8alpha+, human BDCA3+, and sheep CD26+ DCs and is conserved across species. The mRNA encoding the XCR1 ligand chemokine (C motif) ligand 1 (XCL1) is selectively expressed in natural killer (NK) and CD8+ T lymphocytes at steady-state and is enhanced upon activation. Moreover, the Xcl1 mRNA is selectively expressed at high levels in central memory compared with naive CD8+ T lymphocytes. Finally, XCR1-/- mice have decreased early CD8+ T cell responses to Listeria monocytogenes infection, which is associated with higher bacterial loads early in infection. Therefore, XCR1 constitutes the first conserved specific marker for cell subsets homologous to mouse CD8alpha+ DCs in higher vertebrates and promotes their ability to activate early CD8+ T cell defenses against an intracellular pathogenic bacteria.
Figures


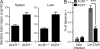
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

