Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo
- PMID: 20479220
- PMCID: PMC2890770
- DOI: 10.1073/pnas.0914507107
Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo
Abstract
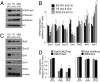
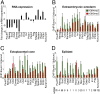
A unique property of the mammalian embryo is that stem cells can be derived from its early tissue lineages. These lineages will give rise to the fetus as well as essential extraembryonic tissues. Understanding how chromatin regulation participates in establishment of these lineages in the embryo and their derived stem cells provides insight that will critically inform our understanding of embryogenesis and stem cell biology. Here, we compare the genomewide location of active and repressive histone modifications in embryonic stem cells, trophoblast stem cells, and extraembryonic endoderm stem cells from the mouse. Our results show that the active modification H3K4me3 has a similar role in the three stem cell types, but the repressive modification H3K27me3 varies in abundance and genomewide distribution. Thus, alternative mechanisms mediate transcriptional repression in stem cells from the embryo. In addition, using carrier chromatin immunoprecipitation we show that bivalent histone domains seen in embryonic stem cells exist in pluripotent cells of the early embryo. However, the epigenetic status of extraembryonic progenitor cells in the embryo did not entirely reflect the extraembryonic stem cell lines. These studies indicate that histone modification mechanisms may differ between early embryo lineages and emphasize the importance of examining in vivo and in vitro progenitor cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

