Evidence that the polymerase of respiratory syncytial virus initiates RNA replication in a nontemplated fashion
- PMID: 20479224
- PMCID: PMC2890450
- DOI: 10.1073/pnas.0913065107
Evidence that the polymerase of respiratory syncytial virus initiates RNA replication in a nontemplated fashion
Abstract
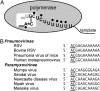
RNA virus polymerases must initiate replicative RNA synthesis with extremely high accuracy to maintain their genome termini and to avoid generating defective genomes. For the single-stranded negative-sense RNA viruses, it is not known how this accuracy is achieved. To investigate this question, mutations were introduced into the 3' terminal base of a respiratory syncytial virus (RSV) template, and the RNA products were examined to determine the impact of the mutation. To perform the assay, RNA replication was reconstituted using a modified minireplicon system in which replication was limited to a single step. Importantly, this system allowed analysis of RSV RNA generated intracellularly, but from a defined template that was not subject to selection by replication. Sequence analysis of RNA products generated from templates containing 1U-C and 1U-A substitutions showed that, in both cases, replication products were initiated with a nontemplated, WT A residue, rather than a templated G or U residue, indicating that the polymerase selects the terminal NTP independently of the template. Examination of a template in which the position 1 nucleotide was deleted supported these findings. This mutant directed efficient replication at approximately 60% of WT levels, and its product was found to be initiated at the WT position (-1 relative to the template) with a WT A residue. These findings show that the RSV replicase selects ATP and initiates at the correct position, independently of the first nucleotide of the template, suggesting a mechanism by which highly accurate replication initiation is achieved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

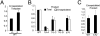

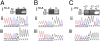
Similar articles
-
The first two nucleotides of the respiratory syncytial virus antigenome RNA replication product can be selected independently of the promoter terminus.RNA. 2011 Oct;17(10):1895-906. doi: 10.1261/rna.2813411. Epub 2011 Aug 30. RNA. 2011. PMID: 21878549 Free PMC article.
-
Mutations in the 5' trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step.J Virol. 2000 Jan;74(1):146-55. doi: 10.1128/jvi.74.1.146-155.2000. J Virol. 2000. PMID: 10590101 Free PMC article.
-
Mechanism for de novo initiation at two sites in the respiratory syncytial virus promoter.Nucleic Acids Res. 2018 Jul 27;46(13):6785-6796. doi: 10.1093/nar/gky480. Nucleic Acids Res. 2018. PMID: 29873775 Free PMC article.
-
How does the polymerase of non-segmented negative strand RNA viruses commit to transcription or genome replication?J Virol. 2024 Aug 20;98(8):e0033224. doi: 10.1128/jvi.00332-24. Epub 2024 Jul 30. J Virol. 2024. PMID: 39078194 Free PMC article. Review.
-
Initiation and regulation of paramyxovirus transcription and replication.Virology. 2015 May;479-480:545-54. doi: 10.1016/j.virol.2015.01.014. Epub 2015 Feb 13. Virology. 2015. PMID: 25683441 Free PMC article. Review.
Cited by
-
Biochemical Effect of Resistance Mutations against Synergistic Inhibitors of RSV RNA Polymerase.PLoS One. 2016 May 10;11(5):e0154097. doi: 10.1371/journal.pone.0154097. eCollection 2016. PLoS One. 2016. PMID: 27163448 Free PMC article.
-
Factors affecting de novo RNA synthesis and back-priming by the respiratory syncytial virus polymerase.Virology. 2014 Aug;462-463:318-27. doi: 10.1016/j.virol.2014.05.032. Epub 2014 Jul 8. Virology. 2014. PMID: 25010481 Free PMC article.
-
Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2'-Fluoro-4'-Chloromethyl-Cytidine Triphosphate.PLoS Pathog. 2015 Jun 22;11(6):e1004995. doi: 10.1371/journal.ppat.1004995. eCollection 2015 Jun. PLoS Pathog. 2015. PMID: 26098424 Free PMC article.
-
Polymerases of paramyxoviruses and pneumoviruses.Virus Res. 2017 Apr 15;234:87-102. doi: 10.1016/j.virusres.2017.01.008. Epub 2017 Jan 16. Virus Res. 2017. PMID: 28104450 Free PMC article. Review.
-
RNA elongation by respiratory syncytial virus polymerase is calibrated by conserved region V.PLoS Pathog. 2017 Dec 27;13(12):e1006803. doi: 10.1371/journal.ppat.1006803. eCollection 2017 Dec. PLoS Pathog. 2017. PMID: 29281742 Free PMC article.
References
-
- van Dijk AA, Makeyev EV, Bamford DH. Initiation of viral RNA-dependent RNA polymerization. J Gen Virol. 2004;85:1077–1093. - PubMed
-
- Paul AV, van Boom JH, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. - PubMed
-
- Hong Z, et al. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology. 2001;285:6–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

