A viral assembly factor promotes AAV2 capsid formation in the nucleolus
- PMID: 20479244
- PMCID: PMC2890453
- DOI: 10.1073/pnas.1001673107
A viral assembly factor promotes AAV2 capsid formation in the nucleolus
Abstract
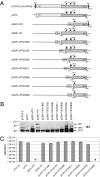
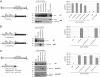
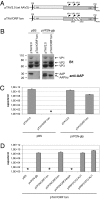
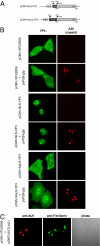
The volume available in icosahedral virus capsids limits the size of viral genomes. To overcome this limitation, viruses have evolved strategies to increase their coding capacity by using more than one ORF while keeping the genome length constant. The assembly of virus capsids requires the coordinated interaction of a large number of subunits to generate a highly ordered structure in which the viral genome can be enclosed. To understand this process, it is essential to know which viral and nonviral components are involved in the assembly reaction. Here, we show that the adeno-associated virus (AAV) encodes a protein required for capsid formation by means of a nested, alternative ORF of the cap gene. Translation is initiated at a nonconventional translation start site, resulting in the expression of a protein with a calculated molecular weight of 23 kDa. This protein, designated assembly-activating protein (AAP), is localized in the host cell nucleolus, where AAV capsid morphogenesis occurs. AAP targets newly synthesized capsid proteins to this organelle and in addition fulfils a function in the assembly reaction itself. Sequence analysis suggests that also all other species of the genus Dependovirus encode a homologous protein in their cap gene. The arrangement of different ORFs that encode capsid proteins and an assembly factor within the same mRNA facilitates a timely coordinated expression of the components involved in the assembly process.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

