Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B
- PMID: 20479468
- PMCID: PMC2872917
- DOI: 10.1083/jcb.200910142
Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B
Abstract
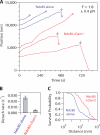
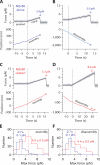
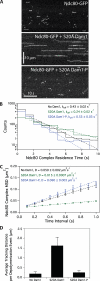
The coupling of kinetochores to dynamic spindle microtubules is crucial for chromosome positioning and segregation, error correction, and cell cycle progression. How these fundamental attachments are made and persist under tensile forces from the spindle remain important questions. As microtubule-binding elements, the budding yeast Ndc80 and Dam1 kinetochore complexes are essential and not redundant, but their distinct contributions are unknown. In this study, we show that the Dam1 complex is a processivity factor for the Ndc80 complex, enhancing the ability of the Ndc80 complex to form load-bearing attachments to and track with dynamic microtubule tips in vitro. Moreover, the interaction between the Ndc80 and Dam1 complexes is abolished when the Dam1 complex is phosphorylated by the yeast aurora B kinase Ipl1. This provides evidence for a mechanism by which aurora B resets aberrant kinetochore-microtubule attachments. We propose that the action of the Dam1 complex as a processivity factor in kinetochore-microtubule attachment is regulated by conserved signals for error correction.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

