Inhibition of hypoxia-inducible factor-1alpha (HIF-1alpha) protein synthesis by DNA damage inducing agents
- PMID: 20479887
- PMCID: PMC2866540
- DOI: 10.1371/journal.pone.0010522
Inhibition of hypoxia-inducible factor-1alpha (HIF-1alpha) protein synthesis by DNA damage inducing agents
Abstract
Hypoxia-inducible factor (HIF) is a heterodimeric transcription factor that is composed of a hypoxia-inducible alpha subunit (HIF-1alpha and HIF-2alpha) and a constitutively expressed beta subunit (HIF-1beta). HIF mediates the adaptation of cells and tissues to low oxygen concentrations. It also plays an important role in tumorigenesis and constitutes an important therapeutic target in anti-tumor therapy. We have screened a number of reported HIF inhibitors for their effects on HIF-transcriptional activity and found that the DNA damage inducing agents camptothecin and mitomycin C produced the most robust effects. Camptothecin is a reported inhibitor of HIF-1alpha translation, while mitomycin C has been reported to induce p53-dependent HIF-1alpha degradation. In this study we demonstrate that the inhibitory effect of mitomycin C on HIF-1alpha protein expression is not dependent on p53 and protein degradation, but also involves HIF-1alpha translational regulation. Initiation of a DNA damage response with the small molecule p53 activator NSC-652287 (RITA) has been reported to inhibit HIF-1alpha protein synthesis by increasing the phosphorylation of eIF2alpha. However, we show here that even when eIF2alpha phosphorylation is prevented, the DNA damage inducing drugs mitomycin C, camptothecin and NSC-652287 still inhibit HIF-1alpha protein synthesis to the same extent. The inhibitory effects of camptothecin on HIF-1alpha expression but not that of mitomycin C and NSC-652287 were dependent on cyclin-dependent kinase activity. In conclusion, specific types of DNA damage can bring about selective inhibition of HIF-1alpha protein synthesis. Further characterization of the involved mechanisms may reveal important novel therapeutic targets.
Conflict of interest statement
Figures


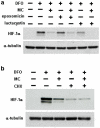


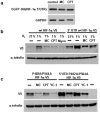
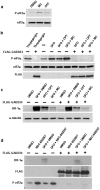
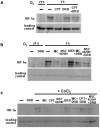
Similar articles
-
Suppression of hypoxia-inducible factor 1α (HIF-1α) by tirapazamine is dependent on eIF2α phosphorylation rather than the mTORC1/4E-BP1 pathway.PLoS One. 2010 Nov 9;5(11):e13910. doi: 10.1371/journal.pone.0013910. PLoS One. 2010. PMID: 21085474 Free PMC article.
-
Chemotherapy-mediated p53-dependent DNA damage response in clear cell renal cell carcinoma: role of the mTORC1/2 and hypoxia-inducible factor pathways.Cell Death Dis. 2013 Oct 17;4(10):e865. doi: 10.1038/cddis.2013.395. Cell Death Dis. 2013. PMID: 24136229 Free PMC article.
-
Small-molecule activation of p53 blocks hypoxia-inducible factor 1alpha and vascular endothelial growth factor expression in vivo and leads to tumor cell apoptosis in normoxia and hypoxia.Mol Cell Biol. 2009 Apr;29(8):2243-53. doi: 10.1128/MCB.00959-08. Epub 2009 Feb 17. Mol Cell Biol. 2009. PMID: 19223463 Free PMC article.
-
Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions.Exp Mol Med. 2004 Feb 29;36(1):1-12. doi: 10.1038/emm.2004.1. Exp Mol Med. 2004. PMID: 15031665 Review.
-
HIF-1alpha and cancer therapy.Recent Results Cancer Res. 2010;180:15-34. doi: 10.1007/978-3-540-78281-0_3. Recent Results Cancer Res. 2010. PMID: 20033376 Review.
Cited by
-
Between Scylla and Charibdis: eIF2α kinases as targets for cancer chemotherapy.Clin Transl Oncol. 2011 Jul;13(7):442-5. doi: 10.1007/s12094-011-0680-3. Clin Transl Oncol. 2011. PMID: 21775270
-
Translational suppression of HIF-1α by miconazole through the mTOR signaling pathway.Cell Oncol (Dordr). 2014 Aug;37(4):269-79. doi: 10.1007/s13402-014-0182-8. Epub 2014 Jul 29. Cell Oncol (Dordr). 2014. PMID: 25070654
-
Similarity in the functions of HIF-1α and HIF-2α proteins in cervical cancer cells.Oncol Lett. 2017 Nov;14(5):5643-5651. doi: 10.3892/ol.2017.6837. Epub 2017 Aug 28. Oncol Lett. 2017. PMID: 29098039 Free PMC article.
-
First-in-human phase 1/2a trial of CRLX101, a cyclodextrin-containing polymer-camptothecin nanopharmaceutical in patients with advanced solid tumor malignancies.Invest New Drugs. 2013 Aug;31(4):986-1000. doi: 10.1007/s10637-012-9921-8. Epub 2013 Feb 9. Invest New Drugs. 2013. PMID: 23397498 Free PMC article. Clinical Trial.
-
Cross-talk between HIF and PD-1/PD-L1 pathways in carcinogenesis and therapy.J Clin Invest. 2022 May 2;132(9):e159473. doi: 10.1172/JCI159473. J Clin Invest. 2022. PMID: 35499071 Free PMC article.
References
-
- Semenza GL. Targeting HIF-1for cancer therapy. Nat Rev Cancer. 2003;3:721–732. - PubMed
-
- Maxwell PH. The HIF pathway in cancer. Semin Cell Dev Biol. 2005;16:523–530. - PubMed
-
- Koshiji M, To KK, Hammer S, Kumamoto K, Harris AL, et al. HIF-1 alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell. 2005;17:793–803. - PubMed
-
- Krishnamachary B, Zagzag D, Nagasawa H, Rainey K, Okuyama H, et al. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel Lindau tumour suppressor-null renal cell carcinoma mediated by TCF, ZFHX1A, and ZFHX1B. Cancer Res. 2006;66:2725–2731. - PubMed
-
- Esteban MA, Tran MG, Harten SK, Hill P, Castellanos MC, et al. Regulation of E-cadherin expression by VHL and hypoxia-inducible factor. Cancer Res. 2006;66:3567–3575. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

