RNA internal loops with tandem AG pairs: the structure of the 5'GAGU/3'UGAG loop can be dramatically different from others, including 5'AAGU/3'UGAA
- PMID: 20481618
- PMCID: PMC2900907
- DOI: 10.1021/bi100332r
RNA internal loops with tandem AG pairs: the structure of the 5'GAGU/3'UGAG loop can be dramatically different from others, including 5'AAGU/3'UGAA
Abstract
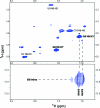
Thermodynamic stabilities of 2 x 2 nucleotide tandem AG internal loops in RNA range from -1.3 to +3.4 kcal/mol at 37 degrees C and are not predicted well with a hydrogen-bonding model. To provide structural information to facilitate development of more sophisticated models for the sequence dependence of stability, we report the NMR solution structures of five RNA duplexes: (rGACGAGCGUCA)(2), (rGACUAGAGUCA)(2), (rGACAAGUGUCA)(2), (rGGUAGGCCA)(2), and (rGACGAGUGUCA)(2). The structures of these duplexes are compared to that of the previously solved (rGGCAGGCC)(2) (Wu, M., SantaLucia, J., Jr., and Turner, D. H. (1997) Biochemistry 36, 4449-4460). For loops bounded by Watson-Crick pairs, the AG and Watson-Crick pairs are all head-to-head imino-paired (cis Watson-Crick/Watson-Crick). The structures suggest that the sequence-dependent stability may reflect non-hydrogen-bonding interactions. Of the two loops bounded by G-U pairs, only the 5'UAGG/3'GGAU loop adopts canonical UG wobble pairing (cis Watson-Crick/Watson-Crick), with AG pairs that are only weakly imino-paired. Strikingly, the 5'GAGU/3'UGAG loop has two distinct duplex conformations, the major of which has both guanosine residues (G4 and G6 in (rGACGAGUGUCA)(2)) in a syn glycosidic bond conformation and forming a sheared GG pair (G4-G6*, GG trans Watson-Crick/Hoogsteen), both uracils (U7 and U7*) flipped out of the helix, and an AA pair (A5-A5*) in a dynamic or stacked conformation. These structures provide benchmarks for computational investigations into interactions responsible for the unexpected differences in loop free energies and structure.
Figures






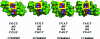
References
-
- Kruger K.; Grabowski P. J.; Zaug A. J.; Sands J.; Gottschling D. E.; Cech T. R. (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31, 147–157. - PubMed
-
- Guerrier-Takada C.; Gardiner K.; Marsh T.; Pace N.; Altman S. (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849–857. - PubMed
-
- Gesteland R. F., Cech T. R., and Atkins J. F. (2006) The RNA World, 3rd ed., Cold Spring Harbor Press, Cold Spring Harbor, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

