Identification of a new 130 bp cis-acting element in the TsVP1 promoter involved in the salt stress response from Thellungiella halophila
- PMID: 20482790
- PMCID: PMC3017807
- DOI: 10.1186/1471-2229-10-90
Identification of a new 130 bp cis-acting element in the TsVP1 promoter involved in the salt stress response from Thellungiella halophila
Abstract
Background: Salt stress is one of the major abiotic stresses affecting plant growth and productivity. Vacuolar H+-pyrophosphatase (H+-PPase) genes play an important role in salt stress tolerance in multiple species.
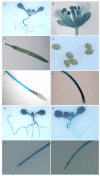
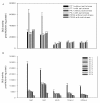
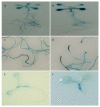
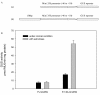
Results: In this study, the promoter from the vacuolar H+-pyrophosphatase from Thellungiella halophila (TsVP1) was cloned and compared with the AVP1 promoter from Arabidopsis thaliana. Sequence analysis indicated that these two promoters had seven similar motifs at similar positions. To determine which tissues the two promoters are active in, transgenic plants were produced with expression of the GUS reporter gene under the control of one of the promoters. In transgenic Arabidopsis with the TsVP1 promoter, the GUS reporter gene had strong activity in almost all tissues except the seeds and the activity was induced in both shoots and roots, especially in the root tips, when treated with salt stress. Such induction was not found in transgenic Arabidopsis with the AVP1 promoter. By analyzing different 5' deletion mutants of the TsVP1 promoter, an 856 bp region (-2200 to -1344) was found to contain enhancer elements that increased gene expression levels. Two AAATGA motifs, which may be the key elements for the anther specific expression profile, in the deleted TsVP1 promoters (PT2 to PT6) were also identified. A 130 bp region (-667 to -538) was finally identified as the key sequence for the salt stress response by analyzing the different mutants both with and without salt stress. GUS transient assay in tobacco leaves suggested the 130 bp region was sufficient for the salt stress response. Bioinformatic analysis also revealed that there may be novel motifs in this region that are the key elements for the salt stress responsive activity of the TsVP1 promoter.
Conclusions: The TsVP1 promoter had strong activity in almost all tissues except the seeds. In addition, its activity was induced by salt stress in leaves and roots, especially in root tips. A 130 bp region (-667 to -538) was identified as the key region for responding to salt stress.
Figures



Similar articles
-
Cloning of an H+-PPase gene from Thellungiella halophila and its heterologous expression to improve tobacco salt tolerance.J Exp Bot. 2006;57(12):3259-70. doi: 10.1093/jxb/erl090. Epub 2006 Aug 28. J Exp Bot. 2006. PMID: 16940040
-
Cloning of a vacuolar H(+)-pyrophosphatase gene from the halophyte Suaeda corniculata whose heterologous overexpression improves salt, saline-alkali and drought tolerance in Arabidopsis.J Integr Plant Biol. 2011 Sep;53(9):731-42. doi: 10.1111/j.1744-7909.2011.01066.x. J Integr Plant Biol. 2011. PMID: 21762382
-
The transcriptional regulatory mechanism of the peroxisomal ascorbate peroxidase (pAPX) gene cloned from an extreme halophyte, Salicornia brachiata.Plant Cell Physiol. 2014 Jan;55(1):201-17. doi: 10.1093/pcp/pct172. Epub 2013 Nov 26. Plant Cell Physiol. 2014. PMID: 24285755
-
Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants.Mol Plant. 2009 Jan;2(1):3-12. doi: 10.1093/mp/ssn094. Mol Plant. 2009. PMID: 19529830 Free PMC article. Review.
-
Plant Promoters: Their Identification, Characterization, and Role in Gene Regulation.Genes (Basel). 2023 Jun 6;14(6):1226. doi: 10.3390/genes14061226. Genes (Basel). 2023. PMID: 37372407 Free PMC article. Review.
Cited by
-
Halophytism: What Have We Learnt From Arabidopsis thaliana Relative Model Systems?Plant Physiol. 2018 Nov;178(3):972-988. doi: 10.1104/pp.18.00863. Epub 2018 Sep 20. Plant Physiol. 2018. PMID: 30237204 Free PMC article. Review.
-
Mechanisms of Plant Responses and Adaptation to Soil Salinity.Innovation (Camb). 2020 Apr 24;1(1):100017. doi: 10.1016/j.xinn.2020.100017. eCollection 2020 May 21. Innovation (Camb). 2020. PMID: 34557705 Free PMC article. Review.
-
Genome-Wide Identification and Salt Stress Response Analysis of the bZIP Transcription Factor Family in Sugar Beet.Int J Mol Sci. 2022 Sep 30;23(19):11573. doi: 10.3390/ijms231911573. Int J Mol Sci. 2022. PMID: 36232881 Free PMC article.
-
Isolation and Functional Validation of Salinity and Osmotic Stress Inducible Promoter from the Maize Type-II H+-Pyrophosphatase Gene by Deletion Analysis in Transgenic Tobacco Plants.PLoS One. 2016 Apr 21;11(4):e0154041. doi: 10.1371/journal.pone.0154041. eCollection 2016. PLoS One. 2016. PMID: 27101137 Free PMC article.
-
Identification of a 467 bp Promoter of Maize Phosphatidylinositol Synthase Gene (ZmPIS) Which Confers High-Level Gene Expression and Salinity or Osmotic Stress Inducibility in Transgenic Tobacco.Front Plant Sci. 2016 Feb 1;7:42. doi: 10.3389/fpls.2016.00042. eCollection 2016. Front Plant Sci. 2016. PMID: 26870063 Free PMC article.
References
-
- Rhoades JD, Loveday J. In: American Society of Agronomists. Steward BA, Nielsen DR, editor. American Society of Civil Engineers, Irrigation of Agricultural Crops; 1990. Salinity in irrigated agriculture; pp. 1089–1142.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

