Proteomics reveals a core molecular response of Pseudomonas putida F1 to acute chromate challenge
- PMID: 20482812
- PMCID: PMC2996968
- DOI: 10.1186/1471-2164-11-311
Proteomics reveals a core molecular response of Pseudomonas putida F1 to acute chromate challenge
Abstract
Background: Pseudomonas putida is a model organism for bioremediation because of its remarkable metabolic versatility, extensive biodegradative functions, and ubiquity in contaminated soil environments. To further the understanding of molecular pathways responding to the heavy metal chromium(VI) [Cr(VI)], the proteome of aerobically grown, Cr(VI)-stressed P. putida strain F1 was characterized within the context of two disparate nutritional environments: rich (LB) media and minimal (M9L) media containing lactate as the sole carbon source.
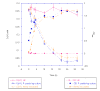
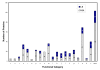
Results: Growth studies demonstrated that F1 sensitivity to Cr(VI) was impacted substantially by nutrient conditions, with a carbon-source-dependent hierarchy (lactate > glucose >> acetate) observed in minimal media. Two-dimensional HPLC-MS/MS was employed to identify differential proteome profiles generated in response to 1 mM chromate under LB and M9L growth conditions. The immediate response to Cr(VI) in LB-grown cells was up-regulation of proteins involved in inorganic ion transport, secondary metabolite biosynthesis and catabolism, and amino acid metabolism. By contrast, the chromate-responsive proteome derived under defined minimal growth conditions was characterized predominantly by up-regulated proteins related to cell envelope biogenesis, inorganic ion transport, and motility. TonB-dependent siderophore receptors involved in ferric iron acquisition and amino acid adenylation domains characterized up-regulated systems under LB-Cr(VI) conditions, while DNA repair proteins and systems scavenging sulfur from alternative sources (e.g., aliphatic sulfonates) tended to predominate the up-regulated proteome profile obtained under M9L-Cr(VI) conditions.
Conclusions: Comparative analysis indicated that the core molecular response to chromate, irrespective of the nutritional conditions tested, comprised seven up-regulated proteins belonging to six different functional categories including transcription, inorganic ion transport/metabolism, and amino acid transport/metabolism. These proteins might potentially serve as indicators of chromate stress in natural microbial communities.
Figures
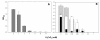

Similar articles
-
Transcriptome analysis reveals response regulator SO2426-mediated gene expression in Shewanella oneidensis MR-1 under chromate challenge.BMC Genomics. 2008 Aug 21;9:395. doi: 10.1186/1471-2164-9-395. BMC Genomics. 2008. PMID: 18718017 Free PMC article.
-
Characterization and genomic analysis of chromate resistant and reducing Bacillus cereus strain SJ1.BMC Microbiol. 2010 Aug 19;10:221. doi: 10.1186/1471-2180-10-221. BMC Microbiol. 2010. PMID: 20723231 Free PMC article.
-
Comparative temporal proteomics of a response regulator (SO2426)-deficient strain and wild-type Shewanella oneidensis MR-1 during chromate transformation.J Proteome Res. 2009 Jan;8(1):59-71. doi: 10.1021/pr800776d. J Proteome Res. 2009. PMID: 19118451
-
The contribution of proteomics to the unveiling of the survival strategies used by Pseudomonas putida in changing and hostile environments.Proteomics. 2013 Oct;13(18-19):2822-30. doi: 10.1002/pmic.201200503. Epub 2013 Jun 18. Proteomics. 2013. PMID: 23625785 Review.
-
Proteomic analysis to unravel the biochemical mechanisms triggered by Bacillus toyonensis SFC 500-1E under chromium(VI) and phenol stress.Biometals. 2023 Oct;36(5):1081-1108. doi: 10.1007/s10534-023-00506-9. Epub 2023 May 20. Biometals. 2023. PMID: 37209221 Review.
Cited by
-
Response to chromate challenge by marine Staphylococcus sp. NIOMR8 evaluated by differential protein expression.3 Biotech. 2018 Dec;8(12):500. doi: 10.1007/s13205-018-1522-6. Epub 2018 Nov 26. 3 Biotech. 2018. PMID: 30498673 Free PMC article.
-
Comparative proteomic analysis reveals mechanistic insights into Pseudomonas putida F1 growth on benzoate and citrate.AMB Express. 2013 Oct 25;3(1):64. doi: 10.1186/2191-0855-3-64. AMB Express. 2013. PMID: 24156539 Free PMC article.
-
Two superoxide dismutases from TnOtchr are involved in detoxification of reactive oxygen species induced by chromate.BMC Microbiol. 2016 Mar 5;16:27. doi: 10.1186/s12866-016-0648-0. BMC Microbiol. 2016. PMID: 26944876 Free PMC article.
-
Engineering sucrose metabolism in Pseudomonas putida highlights the importance of porins.Microb Biotechnol. 2020 Jan;13(1):97-106. doi: 10.1111/1751-7915.13283. Epub 2018 May 28. Microb Biotechnol. 2020. PMID: 29808622 Free PMC article.
-
Extracytoplasmic function (ECF) sigma factor σF is involved in Caulobacter crescentus response to heavy metal stress.BMC Microbiol. 2012 Sep 18;12:210. doi: 10.1186/1471-2180-12-210. BMC Microbiol. 2012. PMID: 22985357 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

