Alzheimer's abeta(1-40) amyloid fibrils feature size-dependent mechanical properties
- PMID: 20483312
- PMCID: PMC2872369
- DOI: 10.1016/j.bpj.2009.12.4317
Alzheimer's abeta(1-40) amyloid fibrils feature size-dependent mechanical properties
Abstract
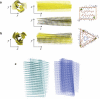
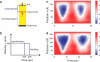
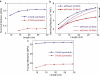
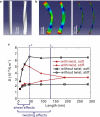
Amyloid fibrils are highly ordered protein aggregates that are associated with several pathological processes, including prion propagation and Alzheimer's disease. A key issue in amyloid science is the need to understand the mechanical properties of amyloid fibrils and fibers to quantify biomechanical interactions with surrounding tissues, and to identify mechanobiological mechanisms associated with changes of material properties as amyloid fibrils grow from nanoscale to microscale structures. Here we report a series of computational studies in which atomistic simulation, elastic network modeling, and finite element simulation are utilized to elucidate the mechanical properties of Alzheimer's Abeta(1-40) amyloid fibrils as a function of the length of the protein filament for both twofold and threefold symmetric amyloid fibrils. We calculate the elastic constants associated with torsional, bending, and tensile deformation as a function of the size of the amyloid fibril, covering fibril lengths ranging from nanometers to micrometers. The resulting Young's moduli are found to be consistent with available experimental measurements obtained from long amyloid fibrils, and predicted to be in the range of 20-31 GPa. Our results show that Abeta(1-40) amyloid fibrils feature a remarkable structural stability and mechanical rigidity for fibrils longer than approximately 100 nm. However, local instabilities that emerge at the ends of short fibrils (on the order of tens of nanometers) reduce their stability and contribute to their disassociation under extreme mechanical or chemical conditions, suggesting that longer amyloid fibrils are more stable. Moreover, we find that amyloids with lengths shorter than the periodicity of their helical pitch, typically between 90 and 130 nm, feature significant size effects of their bending stiffness due the anisotropy in the fibril's cross section. At even smaller lengths (50 nm), shear effects dominate lateral deformation of amyloid fibrils, suggesting that simple Euler-Bernoulli beam models fail to describe the mechanics of amyloid fibrils appropriately. Our studies reveal the importance of size effects in elucidating the mechanical properties of amyloid fibrils. This issue is of great importance for comparing experimental and simulation results, and gaining a general understanding of the biological mechanisms underlying the growth of ectopic amyloid materials.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. - PubMed
-
- Dobson C.M. An overview of protein misfolding diseases. In: Buchner J., Kiefhaber T., editors. Protein Folding Handbook. Wiley-VCH Verlag GMBH & Co. KgaA; Weinheim, Germany: 2005. pp. 1093–1113.
-
- Nelson R., Eisenberg D. Recent atomic models of amyloid fibril structure. Curr. Opin. Struct. Biol. 2006;16:260–265. - PubMed
-
- Harper J.D., Lieber C.M., Lansbury P.T. Atomic force microscopic imaging of seeded fibril formation and fibril branching by the Alzheimer's disease amyloid-β protein. Chem. Biol. 1997;4:951–959. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

