The folding transition-state ensemble of a four-helix bundle protein: helix propensity as a determinant and macromolecular crowding as a probe
- PMID: 20483336
- PMCID: PMC2872220
- DOI: 10.1016/j.bpj.2010.01.052
The folding transition-state ensemble of a four-helix bundle protein: helix propensity as a determinant and macromolecular crowding as a probe
Abstract
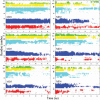
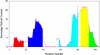
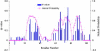
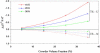
The four-helix bundle protein Rd-apocyt b(562), a redesigned stable variant of apocytochrome b(562), exhibits two-state folding kinetics. Its transition-state ensemble has been characterized by Phi-value analysis. To elucidate the molecular basis of the transition-state ensemble, we have carried out high-temperature molecular dynamics simulations of the unfolding process. In six parallel simulations, unfolding started with the melting of helix I and the C-terminal half of helix IV, and followed by helix III, the N-terminal half of helix IV and helix II. This ordered melting of the helices is consistent with the conclusion from native-state hydrogen exchange, and can be rationalized by differences in intrinsic helix propensity. Guided by experimental Phi-values, a putative transition-state ensemble was extracted from the simulations. The residue helical probabilities of this transition-state ensemble show good correlation with the Phi-values. To further validate the putative transition-state ensemble, the effect of macromolecular crowding on the relative stability between the unfolded ensemble and the transition-state ensemble was calculated. The resulting effect of crowding on the folding kinetics agrees well with experimental observations. This study shows that molecular dynamics simulations combined with calculation of crowding effects provide an avenue for characterize the transition-state ensemble in atomic details.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
An on-pathway hidden intermediate and the early rate-limiting transition state of Rd-apocytochrome b562 characterized by protein engineering.J Mol Biol. 2005 Sep 30;352(4):757-64. doi: 10.1016/j.jmb.2005.07.057. J Mol Biol. 2005. PMID: 16125200
-
Kinetics and motional dynamics of spin-labeled yeast iso-1-cytochrome c: 1. Stopped-flow electron paramagnetic resonance as a probe for protein folding/unfolding of the C-terminal helix spin-labeled at cysteine 102.Biochemistry. 1997 Mar 11;36(10):2884-97. doi: 10.1021/bi962155i. Biochemistry. 1997. PMID: 9062118
-
Amino acid insertion reveals a necessary three-helical intermediate in the folding pathway of the colicin E7 immunity protein Im7.J Mol Biol. 2009 Oct 2;392(4):1074-86. doi: 10.1016/j.jmb.2009.07.085. Epub 2009 Aug 3. J Mol Biol. 2009. PMID: 19651139 Free PMC article.
-
Using simulations to provide the framework for experimental protein folding studies.Arch Biochem Biophys. 2013 Mar;531(1-2):128-35. doi: 10.1016/j.abb.2012.12.015. Epub 2012 Dec 22. Arch Biochem Biophys. 2013. PMID: 23266569 Free PMC article. Review.
-
Principles and Overview of Sampling Methods for Modeling Macromolecular Structure and Dynamics.PLoS Comput Biol. 2016 Apr 28;12(4):e1004619. doi: 10.1371/journal.pcbi.1004619. eCollection 2016 Apr. PLoS Comput Biol. 2016. PMID: 27124275 Free PMC article. Review.
Cited by
-
A method for computing association rate constants of atomistically represented proteins under macromolecular crowding.Phys Biol. 2012 Dec;9(6):066008. doi: 10.1088/1478-3975/9/6/066008. Epub 2012 Nov 29. Phys Biol. 2012. PMID: 23197255 Free PMC article.
-
Quantification of excluded volume effects on the folding landscape of Pseudomonas aeruginosa apoazurin in vitro.Biophys J. 2013 Oct 1;105(7):1689-99. doi: 10.1016/j.bpj.2013.08.038. Biophys J. 2013. PMID: 24094410 Free PMC article.
-
Rate theories for biologists.Q Rev Biophys. 2010 May;43(2):219-93. doi: 10.1017/S0033583510000120. Epub 2010 Aug 9. Q Rev Biophys. 2010. PMID: 20691138 Free PMC article. Review.
-
Simulation and Modeling of Crowding Effects on the Thermodynamic and Kinetic Properties of Proteins with Atomic Details.Biophys Rev. 2013 Jun 1;5(2):207-215. doi: 10.1007/s12551-013-0101-7. Biophys Rev. 2013. PMID: 23710260 Free PMC article.
-
Network representation of conformational transitions between hidden intermediates of Rd-apocytochrome b562.J Chem Phys. 2015 Oct 7;143(13):135101. doi: 10.1063/1.4931921. J Chem Phys. 2015. PMID: 26450332 Free PMC article.
References
-
- Bai Y., Englander S.W. Future directions in folding: the multi-state nature of protein structure. Proteins. 1996;24:145–151. - PubMed
-
- Bai Y. Energy barriers, cooperativity, and hidden intermediates in the folding of small proteins. Biochem. Biophys. Res. Commun. 2006;340:976–983. - PubMed
-
- Chamberlain A.K., Handel T.M., Marqusee S. Detection of rare partially folded molecules in equilibrium with the native conformation of RNaseH. Nat. Struct. Mol. Biol. 1996;3:782–787. - PubMed
-
- Fuentes E.J., Wand A.J. Local stability and dynamics of apocytochrome b562 examined by the dependence of hydrogen exchange on hydrostatic pressure. Biochemistry. 1998;37:9877–9883. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

