Development and application of a comprehensive two-dimensional gas chromatography with time-of-flight mass spectrometry method for the analysis of L-beta-methylamino-alanine in human tissue
- PMID: 20483417
- PMCID: PMC2901168
- DOI: 10.1016/j.chroma.2010.04.065
Development and application of a comprehensive two-dimensional gas chromatography with time-of-flight mass spectrometry method for the analysis of L-beta-methylamino-alanine in human tissue
Abstract
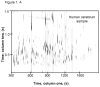
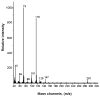
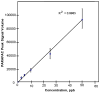
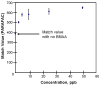
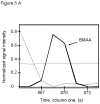
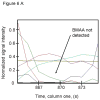
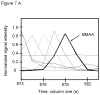
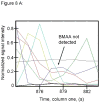
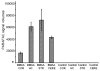
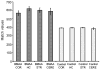
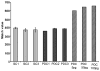
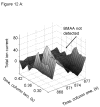
L-Beta-methylamino-alanine (BMAA) has been proposed as a worldwide contributor to neurodegenerative diseases, including Parkinson dementia complex (PDC) of Guam and Alzheimer's disease (AD). Recent conflicting reports of the presence of this amino acid in human brain from patients affected by these diseases have made it necessary to develop methods that provide unambiguous detection in complex samples. Comprehensive two-dimensional gas chromatography coupled with time-of-flight-mass-spectrometry analysis (GCxGC-TOFMS) followed by a targeted Parallel Factor Analysis (PARAFAC) deconvolution method has been used recently in metabolomic investigations to separate, identify, and quantify components of complex biological specimens. We have extended and applied this methodology to the toxicological problem of detecting BMAA in extracts of brain tissue. Our results show that BMAA can be isolated from closely eluting compounds and detected in trace amounts in extracts of brain tissue spiked with low levels of this analyte, ranging from 2.5ppb to 50ppb, with a limit of detection (LOD) of 0.7ppb. This new method was sufficiently sensitive to detect BMAA in cerebral extracts of mice fed BMAA. This optimized approach was then applied to analyze tissue from humans; however, no BMAA was detected in the brain extracts from controls or patients with PDC or AD. Our results demonstrate the application of multidimensional chromatography-mass spectrometry methods and computational deconvolution analysis to the problem of detecting trace amounts of a potential toxin in brain extracts from mice and humans.
Copyright 2010 Elsevier B.V. All rights reserved.
Figures














Similar articles
-
Parkinson-dementia complex and development of a new stable isotope dilution assay for BMAA detection in tissue.Toxicol Appl Pharmacol. 2009 Oct 15;240(2):180-8. doi: 10.1016/j.taap.2009.06.025. Epub 2009 Aug 28. Toxicol Appl Pharmacol. 2009. PMID: 19716838 Free PMC article.
-
The analysis of underivatized β-Methylamino-L-alanine (BMAA), BAMA, AEG & 2,4-DAB in Pteropus mariannus mariannus specimens using HILIC-LC-MS/MS.Toxicon. 2018 Sep 15;152:150-159. doi: 10.1016/j.toxicon.2018.07.028. Epub 2018 Aug 10. Toxicon. 2018. PMID: 30102919 Free PMC article.
-
Matrix Effect of Diverse Biological Samples Extracted with Different Extraction Ratios on the Detection of β-N-Methylamino-L-Alanine by Two Common LC-MS/MS Analysis Methods.Toxins (Basel). 2022 Jun 2;14(6):387. doi: 10.3390/toxins14060387. Toxins (Basel). 2022. PMID: 35737048 Free PMC article.
-
A critical review of the postulated role of the non-essential amino acid, β-N-methylamino-L-alanine, in neurodegenerative disease in humans.J Toxicol Environ Health B Crit Rev. 2017;20(4):1-47. doi: 10.1080/10937404.2017.1297592. Epub 2017 Jun 9. J Toxicol Environ Health B Crit Rev. 2017. PMID: 28598725 Free PMC article. Review.
-
Overview of the potent cyanobacterial neurotoxin β-methylamino-L-alanine (BMAA) and its analytical determination.Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2016 Oct;33(10):1570-1586. doi: 10.1080/19440049.2016.1217070. Epub 2016 Sep 22. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2016. PMID: 27652898 Review.
Cited by
-
Presence of the neurotoxin BMAA in aquatic ecosystems: what do we really know?Toxins (Basel). 2014 Mar 21;6(3):1109-38. doi: 10.3390/toxins6031109. Toxins (Basel). 2014. PMID: 24662480 Free PMC article. Review.
-
The cyanobacteria derived toxin Beta-N-methylamino-L-alanine and amyotrophic lateral sclerosis.Toxins (Basel). 2010 Dec;2(12):2837-50. doi: 10.3390/toxins2122837. Epub 2010 Dec 20. Toxins (Basel). 2010. PMID: 22069578 Free PMC article. Review.
-
Assessment of the non-protein amino acid BMAA in Mediterranean mussel Mytilus galloprovincialis after feeding with estuarine cyanobacteria.Environ Sci Pollut Res Int. 2015 Aug;22(16):12501-10. doi: 10.1007/s11356-015-4516-5. Epub 2015 Apr 24. Environ Sci Pollut Res Int. 2015. PMID: 25903181
-
Forensic significance of VOCs profiling in decayed ante- and post-mortem injuries by GC×GC-TOF/MS.Forensic Sci Med Pathol. 2025 Mar;21(1):138-156. doi: 10.1007/s12024-024-00843-2. Epub 2024 Jul 13. Forensic Sci Med Pathol. 2025. PMID: 39002063
-
β-methylamino-L-alanine (BMAA) is not found in the brains of patients with confirmed Alzheimer's disease.Sci Rep. 2016 Nov 8;6:36363. doi: 10.1038/srep36363. Sci Rep. 2016. PMID: 27821863 Free PMC article.
References
-
- Morris HR, Steele JC, Crook R, Wavrant-De Vrieze F, Onstead-Cardinale L, Gwinn-Hardy K, Wood NW, Farrer M, Lees AJ, McGeer PL, Siddique T, Hardy J, Perez-Tur J. Arch Neurol. 2004;61:1889. - PubMed
-
- Plato CC, Galasko D, Garruto RM, Plato M, Gamst A, Craig UK, Torres JM, Wiederholt W. Neurology. 2002;58:765. - PubMed
-
- Galasko D, Salmon DP, Craig UK, Thal LJ, Schellenberg G, Wiederholt W. Neurology. 2002;58:90. - PubMed
-
- Waring SC. School of Public Health. University of Texas Health Science Center; Houston: 1994.
-
- Vega A, Bell EA. Phytochemistry. 1967;6:759.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

