Vgamma2Vdelta2 T Cell Receptor recognition of prenyl pyrophosphates is dependent on all CDRs
- PMID: 20483784
- PMCID: PMC3069129
- DOI: 10.4049/jimmunol.1000231
Vgamma2Vdelta2 T Cell Receptor recognition of prenyl pyrophosphates is dependent on all CDRs
Abstract
gammadelta T cells differ from alphabeta T cells in the Ags they recognize and their functions in immunity. Although most alphabeta TCRs recognize peptides presented by MHC class I or II, human gammadelta T cells expressing Vgamma2Vdelta2 TCRs recognize nonpeptide prenyl pyrophosphates. To define the molecular basis for this recognition, the effect of mutations in the TCR CDR was assessed. Mutations in all CDR loops altered recognition and cover a large footprint. Unlike murine gammadelta TCR recognition of the MHC class Ib T22 protein, there was no CDR3delta motif required for recognition because only one residue is required. Instead, the length and sequence of CDR3gamma was key. Although a prenyl pyrophosphate-binding site was defined by Lys109 in Jgamma1.2 and Arg51 in CDR2delta, the area outlined by critical mutations is much larger. These results show that prenyl pyrophosphate recognition is primarily by germline-encoded regions of the gammadelta TCR, allowing a high proportion of Vgamma2Vdelta2 TCRs to respond. This underscores its parallels to innate immune receptors. Our results also provide strong evidence for the existence of an Ag-presenting molecule for prenyl pyrophosphates.
Conflict of interest statement
The authors have no conflicting financial interests.
Figures



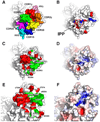
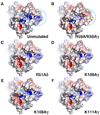

References
-
- Cerundolo V, Silk JD, Masri SH, Salio M. Harnessing invariant NKT cells in vaccination strategies. Nat. Rev. Immunol. 2009;9:28–38. - PubMed
-
- Kronenberg M, Engel I. On the road: progress in finding the unique pathway of invariant NKT cell differentiation. Curr. Opin. Immunol. 2007;19:186–193. - PubMed
-
- Morita CT, Parker CM, Brenner MB, Band H. T cell receptor usage and functional capabilities of human γδ T cells at birth. J. Immunol. 1994;153:3979–3988. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

