A new data mining approach for profiling and categorizing kinetic patterns of metabolic biomarkers after myocardial injury
- PMID: 20483816
- PMCID: PMC2894506
- DOI: 10.1093/bioinformatics/btq254
A new data mining approach for profiling and categorizing kinetic patterns of metabolic biomarkers after myocardial injury
Abstract
Motivation: The discovery of new and unexpected biomarkers in cardiovascular disease is a highly data-driven process that requires the complementary power of modern metabolite profiling technologies, bioinformatics and biostatistics. Clinical biomarkers of early myocardial injury are lacking. A prospective biomarker cohort study was carried out to identify, categorize and profile kinetic patterns of early metabolic biomarkers of planned myocardial infarction (PMI) and spontaneous (SMI) myocardial infarction. We applied a targeted mass spectrometry (MS)-based metabolite profiling platform to serial blood samples drawn from carefully phenotyped patients undergoing alcohol septal ablation for hypertrophic obstructive cardiomyopathy serving as a human model of PMI. Patients with SMI and patients undergoing catheterization without induction of myocardial infarction served as positive and negative controls to assess generalizability of markers identified in PMI.
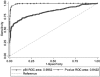
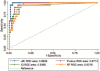
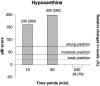
Results: To identify metabolites of high predictive value in tandem mass spectrometry data, we introduced a new feature selection method for the categorization of metabolic signatures into three classes of weak, moderate and strong predictors, which can be easily applied to both paired and unpaired samples. Our paradigm outperformed standard null-hypothesis significance testing and other popular methods for feature selection in terms of the area under the receiver operating curve and the product of sensitivity and specificity. Our results emphasize that this new method was able to identify, classify and validate alterations of levels in multiple metabolites participating in pathways associated with myocardial injury as early as 10 min after PMI.
Availability: The algorithm as well as supplementary material is available for download at: www.umit.at/page.cfm?vpath=departments/technik/iebe/tools/bi
Figures

Similar articles
-
Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury.J Clin Invest. 2008 Oct;118(10):3503-12. doi: 10.1172/JCI35111. J Clin Invest. 2008. PMID: 18769631 Free PMC article. Clinical Trial.
-
Aptamer-Based Proteomic Profiling Reveals Novel Candidate Biomarkers and Pathways in Cardiovascular Disease.Circulation. 2016 Jul 26;134(4):270-85. doi: 10.1161/CIRCULATIONAHA.116.021803. Circulation. 2016. PMID: 27444932 Free PMC article.
-
Application of Large-Scale Aptamer-Based Proteomic Profiling to Planned Myocardial Infarctions.Circulation. 2018 Mar 20;137(12):1270-1277. doi: 10.1161/CIRCULATIONAHA.117.029443. Epub 2017 Dec 8. Circulation. 2018. PMID: 29222138 Free PMC article.
-
Identifying novel biomarkers through data mining-a realistic scenario?Proteomics Clin Appl. 2015 Apr;9(3-4):437-43. doi: 10.1002/prca.201400107. Epub 2015 Jan 12. Proteomics Clin Appl. 2015. PMID: 25347964 Free PMC article. Review.
-
Mass spectrometry-based metabolomics: applications to biomarker and metabolic pathway research.Biomed Chromatogr. 2016 Jan;30(1):7-12. doi: 10.1002/bmc.3453. Epub 2015 Mar 4. Biomed Chromatogr. 2016. PMID: 25739660 Review.
Cited by
-
Prognosis prediction of hepatocellular carcinoma after surgical resection based on serum metabolic profiling from gas chromatography-mass spectrometry.Anal Bioanal Chem. 2021 May;413(12):3153-3165. doi: 10.1007/s00216-021-03281-z. Epub 2021 Apr 2. Anal Bioanal Chem. 2021. PMID: 33796932
-
Prediction of thermostability from amino acid attributes by combination of clustering with attribute weighting: a new vista in engineering enzymes.PLoS One. 2011;6(8):e23146. doi: 10.1371/journal.pone.0023146. Epub 2011 Aug 10. PLoS One. 2011. PMID: 21853079 Free PMC article.
-
Aging-related Changes in Mouse Serum Glycerophospholipid Profiles.Osong Public Health Res Perspect. 2014 Dec;5(6):345-50. doi: 10.1016/j.phrp.2014.10.002. Epub 2014 Nov 13. Osong Public Health Res Perspect. 2014. PMID: 25562043 Free PMC article.
-
Understanding the undelaying mechanism of HA-subtyping in the level of physic-chemical characteristics of protein.PLoS One. 2014 May 8;9(5):e96984. doi: 10.1371/journal.pone.0096984. eCollection 2014. PLoS One. 2014. PMID: 24809455 Free PMC article.
-
Modeling and Classification of Kinetic Patterns of Dynamic Metabolic Biomarkers in Physical Activity.PLoS Comput Biol. 2015 Aug 28;11(8):e1004454. doi: 10.1371/journal.pcbi.1004454. eCollection 2015 Aug. PLoS Comput Biol. 2015. PMID: 26317529 Free PMC article.
References
-
- Ackermann BL, et al. The role of mass spectrometry in biomarker discovery and measurement. Curr. Drug Metab. 2006;7:525–539. - PubMed
-
- Baumgartner C, Baumgartner D. Biomarker discovery, disease classification, and similarity query processing on high-throughput MS/MS data of inborn errors of metabolism. J. Biomol. Screen. 2006;11:90–99. - PubMed
-
- Baumgartner C, Graber A. Data mining and knowledge discovery in metabolomics. In: Masseglia F, Poncelet P, et al., editors. Successes and New Directions in Data Mining. Hershey, PA and London, UK: IGI Global; 2007. pp. 141–166.
-
- Bäckström T, et al. Cardiac outflow of amino acids and purines during myocardial ischemia and reperfusion. J. Appl. Physiol. 2003;94:1122–1128. - PubMed
-
- Collinson PO, Gaze DC. Biomarkers of cardiovascular damage and dysfunction–an overview. Heart Lung Circ. 2007;16(Suppl. 3):S71–S82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

