RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean
- PMID: 20484023
- PMCID: PMC2899914
- DOI: 10.1104/pp.110.158147
RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean
Abstract
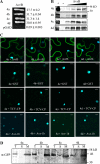
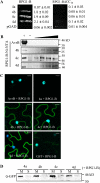
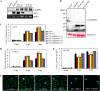
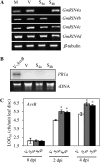
Soybean (Glycine max) RPG1-B (for resistance to Pseudomonas syringae pv glycinea) mediates species-specific resistance to P. syringae expressing the avirulence protein AvrB, similar to the nonorthologous RPM1 in Arabidopsis (Arabidopsis thaliana). RPM1-derived signaling is presumably induced upon AvrB-derived modification of the RPM1-interacting protein, RIN4 (for RPM1-interacting 4). We show that, similar to RPM1, RPG1-B does not directly interact with AvrB but associates with RIN4-like proteins from soybean. Unlike Arabidopsis, soybean contains at least four RIN4-like proteins (GmRIN4a to GmRIN4d). GmRIN4b, but not GmRIN4a, complements the Arabidopsis rin4 mutation. Both GmRIN4a and GmRIN4b bind AvrB, but only GmRIN4b binds RPG1-B. Silencing either GmRIN4a or GmRIN4b abrogates RPG1-B-derived resistance to P. syringae expressing AvrB. Binding studies show that GmRIN4b interacts with GmRIN4a as well as with two other AvrB/RPG1-B-interacting isoforms, GmRIN4c and GmRIN4d. The lack of functional redundancy among GmRIN4a and GmRIN4b and their abilities to interact with each other suggest that the two proteins might function as a heteromeric complex in mediating RPG1-B-derived resistance. Silencing GmRIN4a or GmRIN4b in rpg1-b plants enhances basal resistance to virulent strains of P. syringae and the oomycete Phytophthora sojae. Interestingly, GmRIN4a- or GmRIN4b-silenced rpg1-b plants respond differently to AvrB-expressing bacteria. Although both GmRIN4a and GmRIN4b function to monitor AvrB in the presence of RPG1-B, GmRIN4a, but not GmRIN4b, negatively regulates AvrB virulence activity in the absence of RPG1-B.
Figures


References
-
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE. (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080 - PubMed
-
- Axtell MJ, Staskawicz BJ. (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 - PubMed
-
- Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. (2004) Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16: 2822–2835 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

