Gene positioning
- PMID: 20484389
- PMCID: PMC2869523
- DOI: 10.1101/cshperspect.a000588
Gene positioning
Abstract
Eukaryotic gene expression is an intricate multistep process, regulated within the cell nucleus through the activation or repression of RNA synthesis, processing, cytoplasmic export, and translation into protein. The major regulators of gene expression are chromatin remodeling and transcription machineries that are locally recruited to genes. However, enzymatic activities that act on genes are not ubiquitously distributed throughout the nucleoplasm, but limited to specific and spatially defined foci that promote preferred higher-order chromatin arrangements. The positioning of genes within the nuclear landscape relative to specific functional landmarks plays an important role in gene regulation and disease.
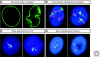
Figures

References
-
- Akhtar A, Gasser SM 2007. The nuclear envelope and transcriptional control. Nat Rev Genet 8:507–517 - PubMed
-
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T 2009. Chromosomal dynamics at the Shh locus: Limb bud-specific differential regulation of competence and active transcription. Dev Cell 16:47–57 - PubMed
-
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394:592–595 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
