Cellular transcription factor ZASC1 regulates murine leukemia virus transcription
- PMID: 20484494
- PMCID: PMC2897642
- DOI: 10.1128/JVI.00299-10
Cellular transcription factor ZASC1 regulates murine leukemia virus transcription
Abstract
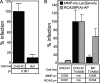
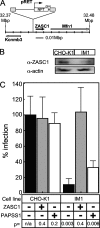
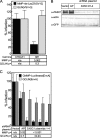
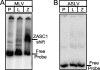
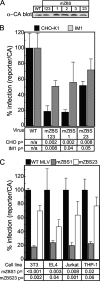
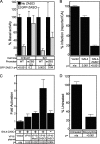
To identify cellular processes involved in retroviral infection, we employed a high-volume forward genetic screen of insertionally mutagenized somatic cells using a murine leukemia virus (MLV) vector. This approach identified a clonal cell line that exhibited approximately 10-fold reduced gene expression from MLV vectors following infection despite supporting normal levels of MLV reverse transcription and integration. The defect in this cell line was specific for the MLV long terminal repeat (LTR) promoter, as normal levels of reporter gene expression were obtained from both an internal cytomegalovirus (CMV) promoter contained within an LTR-defective MLV vector and LTR expression from an avian sarcoma and leukosis virus (ASLV) vector. Complementation and shRNA knockdown experiments demonstrated that the defective gene in these cells is ZASC1 (ZNF639), a transcription factor with strong links to cancer and inherited ataxias. We demonstrated that ZASC1 is a sequence-specific DNA binding protein with three closely related binding sites located within the MLV LTR promoter, but it does not bind to the ASLV promoter. Mutating these putative ZASC1 binding sites significantly reduced levels of MLV gene expression. While wild-type ZASC1 activated expression from the MLV promoter, a green fluorescent protein-ZASC1 fusion protein showed dominant-negative inhibition of MLV gene expression. These studies identify the cellular transcription factor ZASC1 as an activator of MLV gene expression and provide tools that should be useful in studying the links between ZASC1 and human diseases.
Figures



References
-
- Bates, P., J. A. Young, and H. E. Varmus. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043-1051. - PubMed
-
- Bogaerts, S., A. Vanlandschoot, J. van Hengel, and F. van Roy. 2005. Nuclear translocation of alphaN-catenin by the novel zinc finger transcriptional repressor ZASC1. Exp. Cell Res. 311:1-13. - PubMed
-
- Brass, A. L., D. M. Dykxhoorn, Y. Benita, N. Yan, A. Engelman, R. J. Xavier, J. Lieberman, and S. J. Elledge. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921-926. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

