Mutational analysis of the latency-associated nuclear antigen DNA-binding domain of Kaposi's sarcoma-associated herpesvirus reveals structural conservation among gammaherpesvirus origin-binding proteins
- PMID: 20484563
- PMCID: PMC3066550
- DOI: 10.1099/vir.0.020958-0
Mutational analysis of the latency-associated nuclear antigen DNA-binding domain of Kaposi's sarcoma-associated herpesvirus reveals structural conservation among gammaherpesvirus origin-binding proteins
Abstract
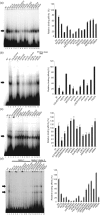
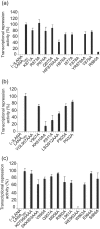
The latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus functions as an origin-binding protein (OBP) and transcriptional regulator. LANA binds the terminal repeats via the C-terminal DNA-binding domain (DBD) to support latent DNA replication. To date, the structure of LANA has not been solved. Sequence alignments among OBPs of gammaherpesviruses have revealed that the C terminus of LANA is structurally related to EBNA1, the OBP of Epstein-Barr virus. Based on secondary structure predictions for LANA(DBD) and published structures of EBNA1(DBD), this study used bioinformatics tools to model a putative structure for LANA(DBD) bound to DNA. To validate the predicted model, 38 mutants targeting the most conserved motifs, namely three alpha-helices and a conserved proline loop, were constructed and functionally tested. In agreement with data for EBNA1, residues in helices 1 and 2 mainly contributed to sequence-specific DNA binding and replication activity, whilst mutations in helix 3 affected replication activity and multimer formation. Additionally, several mutants were isolated with discordant phenotypes, which may aid further studies into LANA function. In summary, these data suggest that the secondary and tertiary structures of LANA and EBNA1 DBDs are conserved and are critical for (i) sequence-specific DNA binding, (ii) multimer formation, (iii) LANA-dependent transcriptional repression, and (iv) DNA replication.
Figures



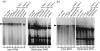
References
-
- Ballestas, M. E., Chatis, P. A. & Kaye, K. M. (1999). Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284, 641–644. - PubMed
-
- Barbera, A. J., Chodaparambil, J. V., Kelley-Clarke, B., Joukov, V., Walter, J. C., Luger, K. & Kaye, K. M. (2006). The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311, 856–861. - PubMed
-
- Bates, P. A., Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. (2001). Enhancement of protein modeling by human intervention in applying the automatic programs 3d-jigsaw and 3d-pssm. Proteins (Suppl. 5), 39–46. - PubMed
-
- Bochkarev, A., Barwell, J. A., Pfuetzner, R. A., Furey, W., Jr, Edwards, A. M. & Frappier, L. (1995). Crystal structure of the DNA-binding domain of the Epstein–Barr virus origin-binding protein EBNA 1. Cell 83, 39–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

