Gata3-deficient mice develop parathyroid abnormalities due to dysregulation of the parathyroid-specific transcription factor Gcm2
- PMID: 20484821
- PMCID: PMC2877956
- DOI: 10.1172/JCI42021
Gata3-deficient mice develop parathyroid abnormalities due to dysregulation of the parathyroid-specific transcription factor Gcm2
Abstract
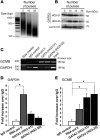
Heterozygous mutations of GATA3, which encodes a dual zinc-finger transcription factor, cause hypoparathyroidism with sensorineural deafness and renal dysplasia. Here, we have investigated the role of GATA3 in parathyroid function by challenging Gata3+/- mice with a diet low in calcium and vitamin D so as to expose any defects in parathyroid function. This led to a higher mortality among Gata3+/- mice compared with Gata3+/+ mice. Compared with their wild-type littermates, Gata3+/- mice had lower plasma concentrations of calcium and parathyroid hormone (PTH) and smaller parathyroid glands with a reduced Ki-67 proliferation rate. At E11.5, Gata3+/- embryos had smaller parathyroid-thymus primordia with fewer cells expressing the parathyroid-specific gene glial cells missing 2 (Gcm2), the homolog of human GCMB. In contrast, E11.5 Gata3-/- embryos had no Gcm2 expression and by E12.5 had gross defects in the third and fourth pharyngeal pouches, including absent parathyroid-thymus primordia. Electrophoretic mobility shift, luciferase reporter, and chromatin immunoprecipitation assays showed that GATA3 binds specifically to a functional double-GATA motif within the GCMB promoter. Thus, GATA3 is critical for the differentiation and survival of parathyroid progenitor cells and, with GCM2/B, forms part of a transcriptional cascade in parathyroid development and function.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

