A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases
- PMID: 20485342
- PMCID: PMC2882514
- DOI: 10.1038/nature08974
A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases
Abstract
Type II topoisomerases are required for the management of DNA tangles and supercoils, and are targets of clinical antibiotics and anti-cancer agents. These enzymes catalyse the ATP-dependent passage of one DNA duplex (the transport or T-segment) through a transient, double-stranded break in another (the gate or G-segment), navigating DNA through the protein using a set of dissociable internal interfaces, or 'gates'. For more than 20 years, it has been established that a pair of dimer-related tyrosines, together with divalent cations, catalyse G-segment cleavage. Recent efforts have proposed that strand scission relies on a 'two-metal mechanism', a ubiquitous biochemical strategy that supports vital cellular processes ranging from DNA synthesis to RNA self-splicing. Here we present the structure of the DNA-binding and cleavage core of Saccharomyces cerevisiae topoisomerase II covalently linked to DNA through its active-site tyrosine at 2.5A resolution, revealing for the first time the organization of a cleavage-competent type II topoisomerase configuration. Unexpectedly, metal-soaking experiments indicate that cleavage is catalysed by a novel variation of the classic two-metal approach. Comparative analyses extend this scheme to explain how distantly-related type IA topoisomerases cleave single-stranded DNA, unifying the cleavage mechanisms for these two essential enzyme families. The structure also highlights a hitherto undiscovered allosteric relay that actuates a molecular 'trapdoor' to prevent subunit dissociation during cleavage. This connection illustrates how an indispensable chromosome-disentangling machine auto-regulates DNA breakage to prevent the aberrant formation of mutagenic and cytotoxic genomic lesions.
Figures

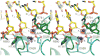

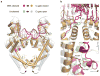
Similar articles
-
Crystal structure of the breakage-reunion domain of DNA gyrase.Nature. 1997 Aug 28;388(6645):903-6. doi: 10.1038/42294. Nature. 1997. PMID: 9278055
-
Similarity in the catalysis of DNA breakage and rejoining by type IA and IIA DNA topoisomerases.Proc Natl Acad Sci U S A. 1999 Feb 2;96(3):881-6. doi: 10.1073/pnas.96.3.881. Proc Natl Acad Sci U S A. 1999. PMID: 9927662 Free PMC article.
-
Structural similarities between topoisomerases that cleave one or both DNA strands.Proc Natl Acad Sci U S A. 1998 Jul 7;95(14):7876-81. doi: 10.1073/pnas.95.14.7876. Proc Natl Acad Sci U S A. 1998. PMID: 9653108 Free PMC article.
-
DNA Topoisomerase Inhibitors: Trapping a DNA-Cleaving Machine in Motion.J Mol Biol. 2019 Aug 23;431(18):3427-3449. doi: 10.1016/j.jmb.2019.07.008. Epub 2019 Jul 10. J Mol Biol. 2019. PMID: 31301408 Free PMC article. Review.
-
What's on the Other Side of the Gate: A Structural Perspective on DNA Gate Opening of Type IA and IIA DNA Topoisomerases.Int J Mol Sci. 2023 Feb 16;24(4):3986. doi: 10.3390/ijms24043986. Int J Mol Sci. 2023. PMID: 36835394 Free PMC article. Review.
Cited by
-
Stereospecific suppression of active site mutants by methylphosphonate substituted substrates reveals the stereochemical course of site-specific DNA recombination.Nucleic Acids Res. 2015 Jul 13;43(12):6023-37. doi: 10.1093/nar/gkv513. Epub 2015 May 20. Nucleic Acids Res. 2015. PMID: 25999343 Free PMC article.
-
Solution structures of DNA-bound gyrase.Nucleic Acids Res. 2011 Jan;39(2):755-66. doi: 10.1093/nar/gkq799. Epub 2010 Sep 24. Nucleic Acids Res. 2011. PMID: 20870749 Free PMC article.
-
Unveiling the interdomain dynamics of type II DNA topoisomerase through all-atom simulations: Implications for understanding its catalytic cycle.Comput Struct Biotechnol J. 2023 Jul 22;21:3746-3759. doi: 10.1016/j.csbj.2023.07.019. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37602233 Free PMC article.
-
Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.Proc Natl Acad Sci U S A. 2016 Feb 16;113(7):1706-13. doi: 10.1073/pnas.1525047113. Epub 2016 Jan 20. Proc Natl Acad Sci U S A. 2016. PMID: 26792525 Free PMC article.
-
Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.Nat Commun. 2015 Dec 7;6:10048. doi: 10.1038/ncomms10048. Nat Commun. 2015. PMID: 26640131 Free PMC article.
References
-
- Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. - PubMed
-
- Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–40. - PubMed
-
- Worland ST, Wang JC. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J Biol Chem. 1989;264:4412–6. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

