22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia
- PMID: 20485365
- PMCID: PMC2977984
- DOI: 10.1038/nrn2841
22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia
Abstract
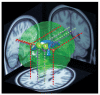
Recent studies are beginning to paint a clear and consistent picture of the impairments in psychological and cognitive competencies that are associated with microdeletions in chromosome 22q11.2. These studies have highlighted a strong link between this genetic lesion and schizophrenia. Parallel studies in humans and animal models are starting to uncover the complex genetic and neural substrates altered by the microdeletion. In addition to offering a deeper understanding of the effects of this genetic lesion, these findings may guide analysis of other copy-number variants associated with cognitive dysfunction and psychiatric disorders.
Figures


References
-
- Shprintzen RJ, et al. A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: velo-cardio-facial syndrome. Cleft Palate J. 1978;15:56–62. - PubMed
-
- DiGeorge A. A new concept of the cellular basis of immunity. Disabil. Rehabil. 1965;67:907–908.
-
- Robin NH, Shprintzen RJ. Defining the clinical spectrum of deletion. 22q11.2. J. Pediatr. 2005;147:90–96. - PubMed
-
- Kobrynski LJ, Sullivan KE. Velocardiofacial syndrome, DiGeorge syndrome: the chromosome 22q11.2 deletion syndromes. Lancet. 2007;370:1443–1452. - PubMed
-
- Botto LD, et al. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics. 2003;112:101–107. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

