Chemical genetics of Plasmodium falciparum
- PMID: 20485428
- PMCID: PMC2874979
- DOI: 10.1038/nature09099
Chemical genetics of Plasmodium falciparum
Abstract
Malaria caused by Plasmodium falciparum is a disease that is responsible for 880,000 deaths per year worldwide. Vaccine development has proved difficult and resistance has emerged for most antimalarial drugs. To discover new antimalarial chemotypes, we have used a phenotypic forward chemical genetic approach to assay 309,474 chemicals. Here we disclose structures and biological activity of the entire library-many of which showed potent in vitro activity against drug-resistant P. falciparum strains-and detailed profiling of 172 representative candidates. A reverse chemical genetic study identified 19 new inhibitors of 4 validated drug targets and 15 novel binders among 61 malarial proteins. Phylochemogenetic profiling in several organisms revealed similarities between Toxoplasma gondii and mammalian cell lines and dissimilarities between P. falciparum and related protozoans. One exemplar compound displayed efficacy in a murine model. Our findings provide the scientific community with new starting points for malaria drug discovery.
Figures
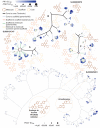
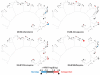

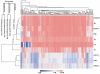
Comment in
-
Drug discovery: Priming the antimalarial pipeline.Nature. 2010 May 20;465(7296):297-8. doi: 10.1038/465297a. Nature. 2010. PMID: 20485420 No abstract available.
-
Antimalarial drugs: A treasure trove of potential antimalarials.Nat Rev Drug Discov. 2010 Jul;9(7):516-7. doi: 10.1038/nrd3209. Nat Rev Drug Discov. 2010. PMID: 20592743 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- AI082617/AI/NIAID NIH HHS/United States
- P01 CA078039/CA/NCI NIH HHS/United States
- AI28724/AI/NIAID NIH HHS/United States
- R01 AI053680/AI/NIAID NIH HHS/United States
- AI075517/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- F32 AI077268/AI/NIAID NIH HHS/United States
- UL1 TR000005/TR/NCATS NIH HHS/United States
- AI053680/AI/NIAID NIH HHS/United States
- P01 AI035707/AI/NIAID NIH HHS/United States
- R21 AI053680/AI/NIAID NIH HHS/United States
- R56 AI053680/AI/NIAID NIH HHS/United States
- R01 AI045774/AI/NIAID NIH HHS/United States
- R56 AI082617/AI/NIAID NIH HHS/United States
- P01 AI067921/AI/NIAID NIH HHS/United States
- AI080625/AI/NIAID NIH HHS/United States
- AI067921/AI/NIAID NIH HHS/United States
- R37 AI028724/AI/NIAID NIH HHS/United States
- CA78039/CA/NCI NIH HHS/United States
- AI772682/AI/NIAID NIH HHS/United States
- U01 AI053862/AI/NIAID NIH HHS/United States
- AI35707/AI/NIAID NIH HHS/United States
- U01 AI075594/AI/NIAID NIH HHS/United States
- P41 RR001614/RR/NCRR NIH HHS/United States
- AI53862/AI/NIAID NIH HHS/United States
- AI075594/AI/NIAID NIH HHS/United States
- AI045774/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

