A large fraction of extragenic RNA pol II transcription sites overlap enhancers
- PMID: 20485488
- PMCID: PMC2867938
- DOI: 10.1371/journal.pbio.1000384
A large fraction of extragenic RNA pol II transcription sites overlap enhancers
Abstract
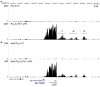
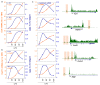
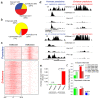
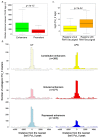
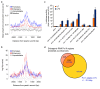
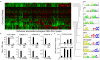
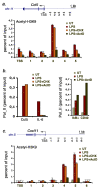
Mammalian genomes are pervasively transcribed outside mapped protein-coding genes. One class of extragenic transcription products is represented by long non-coding RNAs (lncRNAs), some of which result from Pol_II transcription of bona-fide RNA genes. Whether all lncRNAs described insofar are products of RNA genes, however, is still unclear. Here we have characterized transcription sites located outside protein-coding genes in a highly regulated response, macrophage activation by endotoxin. Using chromatin signatures, we could unambiguously classify extragenic Pol_II binding sites as belonging to either canonical RNA genes or transcribed enhancers. Unexpectedly, 70% of extragenic Pol_II peaks were associated with genomic regions with a canonical chromatin signature of enhancers. Enhancer-associated extragenic transcription was frequently adjacent to inducible inflammatory genes, was regulated in response to endotoxin stimulation, and generated very low abundance transcripts. Moreover, transcribed enhancers were under purifying selection and contained binding sites for inflammatory transcription factors, thus suggesting their functionality. These data demonstrate that a large fraction of extragenic Pol_II transcription sites can be ascribed to cis-regulatory genomic regions. Discrimination between lncRNAs generated by canonical RNA genes and products of transcribed enhancers will provide a framework for experimental approaches to lncRNAs and help complete the annotation of mammalian genomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
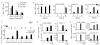
References
-
- Prasanth K. V, Spector D. L. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. - PubMed
-
- Mercer T. R, Dinger M. E, Mattick J. S. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. - PubMed
-
- Sharp P. A. The centrality of RNA. Cell. 2009;136:577–580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

