A comprehensive genome-wide map of autonomously replicating sequences in a naive genome
- PMID: 20485513
- PMCID: PMC2869322
- DOI: 10.1371/journal.pgen.1000946
A comprehensive genome-wide map of autonomously replicating sequences in a naive genome
Abstract
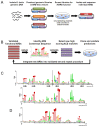
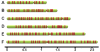
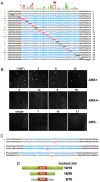
Eukaryotic chromosomes initiate DNA synthesis from multiple replication origins. The machinery that initiates DNA synthesis is highly conserved, but the sites where the replication initiation proteins bind have diverged significantly. Functional comparative genomics is an obvious approach to study the evolution of replication origins. However, to date, the Saccharomyces cerevisiae replication origin map is the only genome map available. Using an iterative approach that combines computational prediction and functional validation, we have generated a high-resolution genome-wide map of DNA replication origins in Kluyveromyces lactis. Unlike other yeasts or metazoans, K. lactis autonomously replicating sequences (KlARSs) contain a 50 bp consensus motif suggestive of a dimeric structure. This motif is necessary and largely sufficient for initiation and was used to dependably identify 145 of the up to 156 non-repetitive intergenic ARSs projected for the K. lactis genome. Though similar in genome sizes, K. lactis has half as many ARSs as its distant relative S. cerevisiae. Comparative genomic analysis shows that ARSs in K. lactis and S. cerevisiae preferentially localize to non-syntenic intergenic regions, linking ARSs with loci of accelerated evolutionary change.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
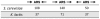

References
-
- Bell S, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

