The type III effectors NleE and NleB from enteropathogenic E. coli and OspZ from Shigella block nuclear translocation of NF-kappaB p65
- PMID: 20485572
- PMCID: PMC2869321
- DOI: 10.1371/journal.ppat.1000898
The type III effectors NleE and NleB from enteropathogenic E. coli and OspZ from Shigella block nuclear translocation of NF-kappaB p65
Abstract
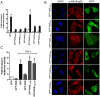
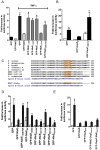
Many bacterial pathogens utilize a type III secretion system to deliver multiple effector proteins into host cells. Here we found that the type III effectors, NleE from enteropathogenic E. coli (EPEC) and OspZ from Shigella, blocked translocation of the p65 subunit of the transcription factor, NF-kappaB, to the host cell nucleus. NF-kappaB inhibition by NleE was associated with decreased IL-8 expression in EPEC-infected intestinal epithelial cells. Ectopically expressed NleE also blocked nuclear translocation of p65 and c-Rel, but not p50 or STAT1/2. NleE homologues from other attaching and effacing pathogens as well OspZ from Shigella flexneri 6 and Shigella boydii, also inhibited NF-kappaB activation and p65 nuclear import; however, a truncated form of OspZ from S. flexneri 2a that carries a 36 amino acid deletion at the C-terminus had no inhibitory activity. We determined that the C-termini of NleE and full length OspZ were functionally interchangeable and identified a six amino acid motif, IDSY(M/I)K, that was important for both NleE- and OspZ-mediated inhibition of NF-kappaB activity. We also established that NleB, encoded directly upstream from NleE, suppressed NF-kappaB activation. Whereas NleE inhibited both TNFalpha and IL-1beta stimulated p65 nuclear translocation and IkappaB degradation, NleB inhibited the TNFalpha pathway only. Neither NleE nor NleB inhibited AP-1 activation, suggesting that the modulatory activity of the effectors was specific for NF-kappaB signaling. Overall our data show that EPEC and Shigella have evolved similar T3SS-dependent means to manipulate host inflammatory pathways by interfering with the activation of selected host transcriptional regulators.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Frankel G, Phillips AD. Attaching effacing Escherichia coli and paradigms of Tir-triggered actin polymerization: getting off the pedestal. Cell Microbiol. 2008;10:549–556. - PubMed
-
- Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, et al. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat Immunol. 2007;8:47–56. - PubMed
-
- Li H, Xu H, Zhou Y, Zhang J, Long C, et al. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–1003. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

