Replication-directed sister chromosome alignment in Escherichia coli
- PMID: 20487299
- PMCID: PMC2859247
- DOI: 10.1111/j.1365-2958.2009.06791.x
Replication-directed sister chromosome alignment in Escherichia coli
Abstract
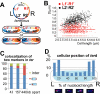
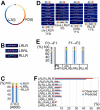
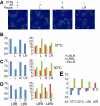
Non-replicating Escherichia coli chromosomes are organized as sausage-shaped structures with the left (L) and the right (R) chromosome arms (replichores) on opposite cell halves and the replication origin (oriC) close to midcell. The replication termination region (ter) therefore passes between the two outer edges of the nucleoid. Four alignment patterns of the two <LR> sister chromosomes within a cell have been detected in an asynchronous population, with the <LRLR> pattern predominating. We test the hypothesis that the minority <LRRL> and <RLLR> patterns arise because of pausing of DNA replication on the right and left replichores respectively. The data resulting from transient pausing or longer-term site-specific blocking of replication show that paused/blocked loci remain close to midcell and the normally replicated-segregated loci locate to the outer regions of the nucleoid, therefore providing experimental support for a direct mechanistic link between DNA replication and chromosome organization.
Figures

References
-
- Espéli O, Mercier R, Boccard F. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol. 2008;68:1418–1427. - PubMed
-
- Lau IF, Filipe SR, Søballe B, Økstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. - PubMed
-
- Mercier R, Petit M-A, Schbath S, Robin S, El-Karoui M, Boccard F, Espéli O. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell. 2008;135:475–485. - PubMed
-
- Michel B, Recchia GD, Penel-Colin M, Ehrlich SD, Sherratt DJ. Resolution of Holliday junctions by RuvABC prevents dimer formation in rep mutants and UV-irradiated cells. Mol Microbiol. 2000;37:180–191. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

