Calmodulin disrupts the structure of the HIV-1 MA protein
- PMID: 20488189
- PMCID: PMC2902600
- DOI: 10.1016/j.jmb.2010.05.022
Calmodulin disrupts the structure of the HIV-1 MA protein
Erratum in
- J Mol Biol. 2011 Oct 28;413(3):742
Abstract
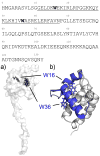
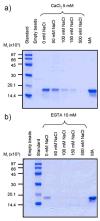
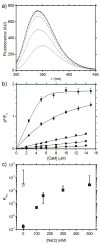
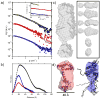
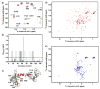
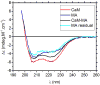
The MA protein from HIV-1 is a small, multifunctional protein responsible for regulating various stages of the viral replication cycle. To achieve its diverse tasks, MA interacts with host cell proteins and it has been reported that one of these is the ubiquitous calcium-sensing calmodulin (CaM), which is up-regulated upon HIV-1 infection. The nature of the CaM-MA interaction has been the subject of structural studies, using peptides based on the MA sequence, that have led to conflicting conclusions. The results presented here show that CaM binds intact MA with 1:1 stoichiometry in a Ca(2+)-dependent manner and that the complex adopts a highly extended conformation in solution as revealed by small-angle X-ray scattering. Alterations in tryptophan fluorescence suggest that the two buried tryptophans (W16 and W36) located in the first two alpha-helices of MA mediate the CaM interaction. Major chemical shift changes occur in the NMR spectrum of MA upon complex formation, whereas chemical shift changes in the CaM spectrum are quite modest and are assigned to residues within the normal target protein-binding hydrophobic clefts of CaM. The NMR data indicate that CaM binds MA via its N- and C-terminal lobes and induces a dramatic conformational change involving a significant loss of secondary and tertiary structure within MA. Circular dichroism experiments suggest that MA loses approximately 20% of its alpha-helical content upon CaM binding. Thus, CaM binding is expected to impact upon the accessibility of interaction sites within MA that are involved in its various functions.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures
References
-
- Radding W, Pan ZQ, Hunter E, Johnston P, Williams JP, McDonald JM. Expression of HIV-1 envelope glycoprotein alters cellular calmodulin. Biochem Biophys Res Commun. 1996;218:192–7. - PubMed
-
- Radding W, Williams JP, McKenna MA, Tummala R, Hunter E, Tytler EM, McDonald JM. Calmodulin and HIV type 1: interactions with Gag and Gag products. AIDS Res Hum Retroviruses. 2000;16:1519–25. - PubMed
-
- Mervis RJ, Ahmad N, Lillehoj EP, Raum MG, Salazar FH, Chan HW, Venkatesan S. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol. 1988;62:3993–4002. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

