Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark
- PMID: 20488795
- PMCID: PMC3222143
- DOI: 10.4049/jimmunol.0902774
Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark
Abstract
Cartilaginous fish are the oldest animals that generate RAG-based Ag receptor diversity. We have analyzed the genes and expressed transcripts of the four TCR chains for the first time in a cartilaginous fish, the nurse shark (Ginglymostoma cirratum). Northern blotting found TCR mRNA expression predominantly in lymphoid and mucosal tissues. Southern blotting suggested translocon-type loci encoding all four chains. Based on diversity of V and J segments, the expressed combinatorial diversity for gamma is similar to that of human, alpha and beta may be slightly lower, and delta diversity is the highest of any organism studied to date. Nurse shark TCRdelta have long CDR3 loops compared with the other three chains, creating binding site topologies comparable to those of mammalian TCR in basic paratope structure; additionally, nurse shark TCRdelta CDR3 are more similar to IgH CDR3 in length and heterogeneity than to other TCR chains. Most interestingly, several cDNAs were isolated that contained IgM or IgW V segments rearranged to other gene segments of TCRdelta and alpha. Finally, in situ hybridization experiments demonstrate a conservation of both alpha/beta and gamma/delta T cell localization in the thymus across 450 million years of vertebrate evolution, with gamma/delta TCR expression especially high in the subcapsular region. Collectively, these data make the first cellular identification of TCR-expressing lymphocytes in a cartilaginous fish.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures

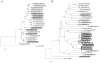



Similar articles
-
Ancient Use of Ig Variable Domains Contributes Significantly to the TCRδ Repertoire.J Immunol. 2019 Sep 1;203(5):1265-1275. doi: 10.4049/jimmunol.1900369. Epub 2019 Jul 24. J Immunol. 2019. PMID: 31341077 Free PMC article.
-
Nurse shark T-cell receptors employ somatic hypermutation preferentially to alter alpha/delta variable segments associated with alpha constant region.Eur J Immunol. 2020 Sep;50(9):1307-1320. doi: 10.1002/eji.201948495. Epub 2020 Jun 17. Eur J Immunol. 2020. PMID: 32346855 Free PMC article.
-
Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark.BMC Immunol. 2004 May 6;5:8. doi: 10.1186/1471-2172-5-8. BMC Immunol. 2004. PMID: 15132758 Free PMC article.
-
Lost structural and functional inter-relationships between Ig and TCR loci in mammals revealed in sharks.Immunogenetics. 2021 Feb;73(1):17-33. doi: 10.1007/s00251-020-01183-5. Epub 2021 Jan 15. Immunogenetics. 2021. PMID: 33449123 Free PMC article. Review.
-
T-cell receptors in ectothermic vertebrates.Immunol Rev. 1998 Dec;166:87-102. doi: 10.1111/j.1600-065x.1998.tb01255.x. Immunol Rev. 1998. PMID: 9914905 Review.
Cited by
-
A Comparison of the Innate and Adaptive Immune Systems in Cartilaginous Fish, Ray-Finned Fish, and Lobe-Finned Fish.Front Immunol. 2019 Oct 10;10:2292. doi: 10.3389/fimmu.2019.02292. eCollection 2019. Front Immunol. 2019. PMID: 31649660 Free PMC article. Review.
-
A conserved αβ transmembrane interface forms the core of a compact T-cell receptor-CD3 structure within the membrane.Proc Natl Acad Sci U S A. 2016 Oct 25;113(43):E6649-E6658. doi: 10.1073/pnas.1611445113. Epub 2016 Oct 10. Proc Natl Acad Sci U S A. 2016. PMID: 27791034 Free PMC article.
-
Shark IgW C region diversification through RNA processing and isotype switching.J Immunol. 2013 Sep 15;191(6):3410-8. doi: 10.4049/jimmunol.1301257. Epub 2013 Aug 9. J Immunol. 2013. PMID: 23935192 Free PMC article.
-
The first crystal structure of CD8αα from a cartilaginous fish.Front Immunol. 2023 Apr 14;14:1156219. doi: 10.3389/fimmu.2023.1156219. eCollection 2023. Front Immunol. 2023. PMID: 37122697 Free PMC article.
-
"Double-duty" conventional dendritic cells in the amphibian Xenopus as the prototype for antigen presentation to B cells.Eur J Immunol. 2018 Mar;48(3):430-440. doi: 10.1002/eji.201747260. Epub 2018 Feb 14. Eur J Immunol. 2018. PMID: 29235109 Free PMC article.
References
-
- Flajnik MF, Rumfelt LL. The immune system of cartilaginous fish. Curr Top Microbiol Immunol. 2000;248:249–270. - PubMed
-
- Hinds KR, Litman GW. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature. 1986;320:546–549. - PubMed
-
- Eason DD, Litman RT, Luer CA, Kerr W, Litman GW. Expression of individual immunoglobulin genes occurs in an unusual system consisting of multiple independent loci. Eur J Immunol. 2004;34:2551–2558. - PubMed
-
- Rast JP, Anderson MK, Ota T, Litman RT, Margittai M, Shamblott MJ, Litman GW. Immunoglobulin light chain class multiplicity and alternative organizational forms in early vertebrate phylogeny. Immunogenetics. 1994;40:83–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

