Sequence conversion by single strand oligonucleotide donors via non-homologous end joining in mammalian cells
- PMID: 20489199
- PMCID: PMC2906313
- DOI: 10.1074/jbc.M110.123844
Sequence conversion by single strand oligonucleotide donors via non-homologous end joining in mammalian cells
Abstract
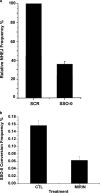
Double strand breaks (DSBs) can be repaired by homology independent nonhomologous end joining (NHEJ) pathways involving proteins such as Ku70/80, DNAPKcs, Xrcc4/Ligase 4, and the Mre11/Rad50/Nbs1 (MRN) complex. DSBs can also be repaired by homology-dependent pathways (HDR), in which the MRN and CtIP nucleases produce single strand ends that engage homologous sequences either by strand invasion or strand annealing. The entry of ends into HDR pathways underlies protocols for genomic manipulation that combine site-specific DSBs with appropriate informational donors. Most strategies utilize long duplex donors that participate by strand invasion. Work in yeast indicates that single strand oligonucleotide (SSO) donors are also active, over considerable distance, via a single strand annealing pathway. We examined the activity of SSO donors in mammalian cells at DSBs induced either by a restriction nuclease or by a targeted interstrand cross-link. SSO donors were effective immediately adjacent to the break, but activity declined sharply beyond approximately 100 nucleotides. Overexpression of the resection nuclease CtIP increased the frequency of SSO-mediated sequence modulation distal to the break site, but had no effect on the activity of an SSO donor adjacent to the break. Genetic and in vivo competition experiments showed that sequence conversion by SSOs in the immediate vicinity of the break was not by strand invasion or strand annealing pathways. Instead these donors competed for ends that would have otherwise entered NHEJ pathways.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

