Decrease in membrane phospholipid unsaturation induces unfolded protein response
- PMID: 20489212
- PMCID: PMC2903364
- DOI: 10.1074/jbc.M110.126870
Decrease in membrane phospholipid unsaturation induces unfolded protein response
Abstract
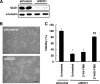
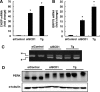
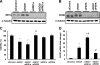
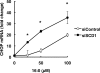
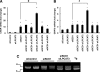
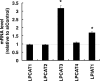
Various kinds of fatty acids are distributed in membrane phospholipids in mammalian cells and tissues. The degree of fatty acid unsaturation in membrane phospholipids affects many membrane-associated functions and can be influenced by diet and by altered activities of lipid-metabolizing enzymes such as fatty acid desaturases. However, little is known about how mammalian cells respond to changes in phospholipid fatty acid composition. In this study we showed that stearoyl-CoA desaturase 1 (SCD1) knockdown increased the amount of saturated fatty acids and decreased that of monounsaturated fatty acids in phospholipids without affecting the amount or the composition of free fatty acid and induced unfolded protein response (UPR), evidenced by increased expression of C/EBP homologous protein (CHOP) and glucose-regulated protein 78 (GRP78) mRNAs and splicing of Xbox-binding protein 1 (XBP1) mRNA. SCD1 knockdown-induced UPR was rescued by various unsaturated fatty acids and was enhanced by saturated fatty acid. Lysophosphatidylcholine acyltransferase 3 (LPCAT3), which incorporates preferentially polyunsaturated fatty acids into phosphatidylcholine, was up-regulated in SCD1 knockdown cells. Knockdown of LPCAT3 synergistically enhanced UPR with SCD1 knockdown. Finally we showed that palmitic acid-induced UPR was significantly enhanced by LPCAT3 knockdown as well as SCD1 knockdown. These results suggest that a decrease in membrane phospholipid unsaturation induces UPR.
Figures


References
-
- Lands W. E., Crawford C. G. (1976) Enzymes of Membrane Phospholipid Metabolism in Animals, 2nd Ed., pp. 3–85, Plenum Press, New York
-
- Holub B. J., Kuksis A. (1978) Adv. Lipid Res 16, 1–125 - PubMed
-
- MacDonald J. I., Sprecher H. (1991) Biochim. Biophys. Acta 1084, 105–121 - PubMed
-
- Spector A. A., Yorek M. A. (1985) J. Lipid Res. 26, 1015–1035 - PubMed
-
- Clandinin M. T., Cheema S., Field C. J., Garg M. L., Venkatraman J., Clandinin T. R. (1991) FASEB J. 5, 2761–2769 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

