Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens
- PMID: 20492702
- PMCID: PMC2901363
- DOI: 10.1186/1475-2891-9-23
Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens
Abstract
Background: The influence of diet on intestinal microflora has been investigated mainly using conventional microbiological approaches. Although these studies have advanced knowledge on human intestinal microflora, it is imperative that new methods are applied to facilitate scientific progress. Culture-independent molecular fingerprinting method of Polymerase Chain Reaction and Denaturing Gradient Gel Electrophoresis (PCR-DGGE) has been used to study microbial communities in a variety of environmental samples. However, these protocols must be optimized prior to their application in order to enhance the quality and accuracy of downstream analyses. In this study, the relative efficacy of four commercial DNA extraction kits (Mobio Ultra Clean(R) Fecal DNA Isolation Kit, M; QIAamp DNA Stool Mini Kit, Q; FastDNA SPIN Kit, FSp; FastDNA SPIN Kit for Soil, FSo) were evaluated. Further, PCR-DGGE technique was also assessed for its feasibility in detecting differences in human intestinal bacterial fingerprint profiles.
Method: Total DNA was extracted from varying weights of human fecal specimens using four different kits, followed by PCR amplification of bacterial 16S rRNA genes, and DGGE separation of the amplicons.
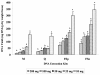
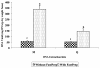
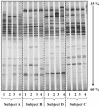
Results: Regardless of kit, maximum DNA yield was obtained using 10 to 50 mg (wet wt) of fecal specimens and similar DGGE profiles were obtained. However, kits FSp and FSo extracted significantly larger amounts of DNA per g dry fecal specimens and produced more bands on their DGGE profiles than kits M and Q due to their use of bead-containing lysing matrix and vigorous shaking step. DGGE of 16S rRNA gene PCR products was suitable for capturing the profiles of human intestinal microbial community and enabled rapid comparative assessment of inter- and intra-subject differences.
Conclusion: We conclude that extraction kits that incorporated bead-containing lysing matrix and vigorous shaking produced high quality DNA from human fecal specimens (10 to 50 mg, wet wt) that can be resolved as bacterial community fingerprints using PCR-DGGE technique. Subsequently, PCR-DGGE technique can be applied for studying variations in human intestinal microbial communities.
Figures



References
-
- Whitney E, Rolfes S. Understanding Nutrition. 9. Wadsworth Thomson Learning; 2002. The fat soluble vitamins: A, D, E, and K; pp. 370–372.
-
- Tannock GW. Normal microflora: an introduction to microbes inhabiting the human body. Chapman & Hall; 1995. Invisible forces: the influence of the normal microflora on host characteristics; pp. 63–84.
-
- Rastall R, Gibson G, Gill H, Guarner F, Klaenhammer T, Pot B, Reid G, Rowland I, Sanders M. Modulation of the microbial ecology of the human colon by probiotics, prebiotics and synbiotics to enhance human health: an overview of enabling science and potential applications. FEMS Microbiol Ecol. 2005;52(2):145–152. doi: 10.1016/j.femsec.2005.01.003. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

