Characterization of the GXXXG motif in the first transmembrane segment of Japanese encephalitis virus precursor membrane (prM) protein
- PMID: 20492732
- PMCID: PMC2890656
- DOI: 10.1186/1423-0127-17-39
Characterization of the GXXXG motif in the first transmembrane segment of Japanese encephalitis virus precursor membrane (prM) protein
Abstract
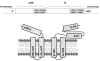
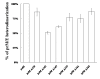
The interaction between prM and E proteins in flavivirus-infected cells is a major driving force for the assembly of flavivirus particles. We used site-directed mutagenesis to study the potential role of the transmembrane domains of the prM proteins of Japanese encephalitis virus (JEV) in prM-E heterodimerization as well as subviral particle formation. Alanine insertion scanning mutagenesis within the GXXXG motif in the first transmembrane segment of JEV prM protein affected the prM-E heterodimerization; its specificity was confirmed by replacing the two glycines of the GXXXG motif with alanine, leucine and valine. The GXXXG motif was found to be conserved in the JEV serocomplex viruses but not other flavivirus groups. These mutants with alanine inserted in the two prM transmembrane segments all impaired subviral particle formation in cell cultures. The prM transmembrane domains of JEV may play importation roles in prM-E heterodimerization and viral particle assembly.
Figures








Similar articles
-
Glutamic acid at residue 125 of the prM helix domain interacts with positively charged amino acids in E protein domain II for Japanese encephalitis virus-like-particle production.J Virol. 2014 Aug;88(15):8386-96. doi: 10.1128/JVI.00937-14. Epub 2014 May 14. J Virol. 2014. PMID: 24829339 Free PMC article.
-
Histidine at residue 99 and the transmembrane region of the precursor membrane prM protein are important for the prM-E heterodimeric complex formation of Japanese encephalitis virus.J Virol. 2005 Jul;79(13):8535-44. doi: 10.1128/JVI.79.13.8535-8544.2005. J Virol. 2005. PMID: 15956595 Free PMC article.
-
Role of the transmembrane domains of prM and E proteins in the formation of yellow fever virus envelope.J Virol. 2003 Jan;77(2):813-20. doi: 10.1128/jvi.77.2.813-820.2003. J Virol. 2003. PMID: 12502797 Free PMC article.
-
A conserved region in the prM protein is a critical determinant in the assembly of flavivirus particles.J Gen Virol. 2012 Jan;93(Pt 1):27-38. doi: 10.1099/vir.0.035964-0. Epub 2011 Sep 28. J Gen Virol. 2012. PMID: 21957123
-
Virus evolution: how does an enveloped virus make a regular structure?Cell. 2001 Apr 6;105(1):5-8. doi: 10.1016/s0092-8674(01)00291-4. Cell. 2001. PMID: 11300997 Free PMC article. Review. No abstract available.
Cited by
-
Directional entry and release of Zika virus from polarized epithelial cells.Virol J. 2019 Aug 8;16(1):99. doi: 10.1186/s12985-019-1200-2. Virol J. 2019. PMID: 31395061 Free PMC article.
-
Protein oligomerization mediated by the transmembrane carboxyl terminal domain of Bcl-XL.FEBS Lett. 2011 Oct 3;585(19):2935-42. doi: 10.1016/j.febslet.2011.08.012. Epub 2011 Aug 16. FEBS Lett. 2011. PMID: 21856303 Free PMC article.
-
A Single Amino Acid Substitution in the M Protein Attenuates Japanese Encephalitis Virus in Mammalian Hosts.J Virol. 2015 Dec 9;90(5):2676-89. doi: 10.1128/JVI.01176-15. J Virol. 2015. PMID: 26656690 Free PMC article.
-
A Molecular Determinant of West Nile Virus Secretion and Morphology as a Target for Viral Attenuation.J Virol. 2020 Jun 1;94(12):e00086-20. doi: 10.1128/JVI.00086-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32269117 Free PMC article.
-
Transmembrane helix dimerization: beyond the search for sequence motifs.Biochim Biophys Acta. 2012 Feb;1818(2):183-93. doi: 10.1016/j.bbamem.2011.08.031. Epub 2011 Sep 1. Biochim Biophys Acta. 2012. PMID: 21910966 Free PMC article. Review.
References
-
- Burke D. In: Fields virology. 4. Knipe DM, Howley P, editor. 2001. Flaviviruses; pp. 1043–1125.
-
- Linderbach B. In: Fields virology, Lippincott-Raven, Philadelphia, Pa. 4. Knipe DM, Howley P, editor. 2001. Flaviviridae: the viruses and their replication; pp. 991–1041.
-
- Russel P. The Togaviruses. Academic Press, New York, NY; 1980. pp. 503–529.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

