beta-hairpin-forming peptides; models of early stages of protein folding
- PMID: 20494507
- PMCID: PMC2906654
- DOI: 10.1016/j.bpc.2010.05.001
beta-hairpin-forming peptides; models of early stages of protein folding
Abstract
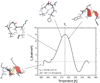
Formation of beta-hairpins is considered the initial step of folding of many proteins and, consequently, peptides constituting the beta-hairpin sequence of proteins (the beta-hairpin-forming peptides) are considered as models of early stages of protein folding. In this article, we discuss the results of experimental studies (circular-dichroism, infrared and nuclear magnetic resonance spectroscopy, and differential scanning calorimetry) of the structure of beta-hairpin-forming peptides excised from the B1 domain of protein G, which are known to fold on their own. We demonstrate that local interactions at the turn sequence and hydrophobic interactions between nonpolar residues are the dominant structure-determining factors, while there is no convincing evidence that stable backbone hydrogen bonds are formed in these peptides in aqueous solution. Consequently, the most plausible mechanism for folding of the beta-hairpin sequence appears to be the broken-zipper mechanism consisting of the following three steps: (i) bending the chain at the turn sequence owing to favorable local interactions, (ii) formation of loose hydrophobic contacts between nonpolar residues, which occur close to the contacts in the native structure of the protein but not exactly in the same position and, finally, (iii) formation of backbone hydrogen bonds and locking the hydrophobic contacts in the native positions as a hydrophobic core develops, sufficient to dehydrate the backbone peptide groups. This mechanism provides sufficient uniqueness (contacts form between residues that become close together because the chain is bent at the turn position) and robustness (contacts need not occur at once in the native positions) for folding a beta-hairpin sequence.
2010 Elsevier B.V. All rights reserved.
Figures



Similar articles
-
Mechanism of formation of the C-terminal beta-hairpin of the B3 domain of the immunoglobulin binding protein G from Streptococcus. I. Importance of hydrophobic interactions in stabilization of beta-hairpin structure.Proteins. 2009 Jun;75(4):931-53. doi: 10.1002/prot.22304. Proteins. 2009. PMID: 19089955 Free PMC article.
-
Dissecting the stability of a beta-hairpin peptide that folds in water: NMR and molecular dynamics analysis of the beta-turn and beta-strand contributions to folding.J Mol Biol. 1999 Oct 8;292(5):1051-69. doi: 10.1006/jmbi.1999.3119. J Mol Biol. 1999. PMID: 10512702
-
Folding dynamics of 10-residue beta-hairpin peptide chignolin.Chem Asian J. 2007 May 4;2(5):591-8. doi: 10.1002/asia.200600385. Chem Asian J. 2007. PMID: 17465405
-
Infrared study of the stability and folding kinetics of a series of β-hairpin peptides with a common NPDG turn.J Phys Chem B. 2011 Dec 29;115(51):15332-8. doi: 10.1021/jp2046867. Epub 2011 Dec 2. J Phys Chem B. 2011. PMID: 22136248 Free PMC article.
-
Roles of beta-turns in protein folding: from peptide models to protein engineering.Biopolymers. 2008 May;89(5):380-91. doi: 10.1002/bip.20960. Biopolymers. 2008. PMID: 18275088 Free PMC article. Review.
Cited by
-
Improvement of the treatment of loop structures in the UNRES force field by inclusion of coupling between backbone- and side-chain-local conformational states.J Chem Theory Comput. 2013 Oct 8;9(10):10.1021/ct4004977. doi: 10.1021/ct4004977. J Chem Theory Comput. 2013. PMID: 24273465 Free PMC article.
-
Folding of Truncated Granulin Peptides.Biomolecules. 2020 Aug 6;10(8):1152. doi: 10.3390/biom10081152. Biomolecules. 2020. PMID: 32781704 Free PMC article.
-
Mutation-induced change in chignolin stability from π-turn to α-turn.RSC Adv. 2020 Jun 15;10(38):22797-22808. doi: 10.1039/d0ra01148g. eCollection 2020 Jun 10. RSC Adv. 2020. PMID: 35514567 Free PMC article.
-
Kinks, loops, and protein folding, with protein A as an example.J Chem Phys. 2014 Jan 14;140(2):025101. doi: 10.1063/1.4855735. J Chem Phys. 2014. PMID: 24437917 Free PMC article.
-
Deceleration of arginine kinase refolding by induced helical structures.Protein J. 2012 Apr;31(4):267-74. doi: 10.1007/s10930-012-9397-6. Protein J. 2012. PMID: 22407434
References
-
- Cox JPL, Evans PA, Packman LC, Williams DH, Woolfson DN. Dissecting the structure of a partially folded protein: circular dichroism and nuclear magnetic resonance studies of peptides from ubiquitin. J. Mol. Biol. 1993;234:483–492. - PubMed
-
- Blanco FJ, Jiménez MA, Herranz J, Rico M, Santoro J, Nieto J. NMR evidence of a short linear peptide that folds into a β-hairpin in aqueous solution. J. Am. Chem. Soc. 1993;115:5887–5888.
-
- Blanco FJ, Rivas FG, Serrano L. A short linear peptide that folds into a stable β-hairpin in aqueous solution. Nature Struct. Biol. 1994;1:584–590. - PubMed
-
- Searle MS, Williams DH, Packman LC. A short linear peptide derived from the N-terminal sequence of ubiquitin folds into a water-stable β-hairpin. Nature Struct. Biol. 1995;2:999–1006. - PubMed
-
- Cline LL, Waters ML. Design of a β-hairpin peptide-intercalator conjugate for simultaneous recognition of single stranded and double stranded regions of RNA. Organic & Biomol. Chem. 2009;7:4622–4630. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

