A sequence that affects the copy number and stability of pSW200 and ColE1
- PMID: 20494993
- PMCID: PMC2897340
- DOI: 10.1128/JB.00095-10
A sequence that affects the copy number and stability of pSW200 and ColE1
Abstract
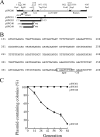
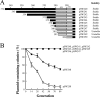
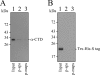
Pantoea stewartii SW2 contains 13 plasmids. One of these plasmids, pSW200, has a replicon that resembles that of ColE1. This study demonstrates that pSW200 contains a 9-bp UP element, 5'-AAGATCTTC, which is located immediately upstream of the -35 box in the RNAII promoter. A transcriptional fusion study reveals that substituting this 9-bp sequence reduces the activity of the RNAII promoter by 78%. The same mutation also reduced the number of plasmid copies from 13 to 5, as well as the plasmid stability. When a similar sequence in a ColE1 derivative, pYCW301, is mutated, the copy number of the plasmid also declines from 34 to 16 per cell. Additionally, inserting this 9-bp sequence stabilizes an unstable pSW100 derivative, pSW142K, which also contains a replicon resembling that of ColE1, indicating the importance of this sequence in maintaining the stability of the plasmid. In conclusion, the 9-bp sequence upstream of the -35 box in the RNAII promoter is required for the efficient synthesis of RNAII and maintenance of the stability of the plasmids in the ColE1 family.
Figures



Similar articles
-
A repeat sequence causes competition of ColE1-type plasmids.PLoS One. 2013 Apr 16;8(4):e61668. doi: 10.1371/journal.pone.0061668. Print 2013. PLoS One. 2013. PMID: 23613898 Free PMC article.
-
Stabilization of pSW100 from Pantoea stewartii by the F conjugation system.J Bacteriol. 2008 May;190(10):3681-9. doi: 10.1128/JB.00846-07. Epub 2008 Mar 14. J Bacteriol. 2008. PMID: 18344358 Free PMC article.
-
Sequence analysis of an Erwinia stewartii plasmid, pSW100.Plasmid. 1995 Sep;34(2):75-84. doi: 10.1006/plas.1995.9999. Plasmid. 1995. PMID: 8559805
-
Isolation and characterization of plasmid pSW200 from Erwinia stewartii.Plasmid. 1998 Sep;40(2):100-12. doi: 10.1006/plas.1998.1350. Plasmid. 1998. PMID: 9735312
-
RNAII transcribed by IPTG-induced T7 RNA polymerase is non-functional as a replication primer for ColE1-type plasmids in Escherichia coli.Nucleic Acids Res. 1995 May 25;23(10):1691-5. doi: 10.1093/nar/23.10.1691. Nucleic Acids Res. 1995. PMID: 7540285 Free PMC article.
Cited by
-
A repeat sequence causes competition of ColE1-type plasmids.PLoS One. 2013 Apr 16;8(4):e61668. doi: 10.1371/journal.pone.0061668. Print 2013. PLoS One. 2013. PMID: 23613898 Free PMC article.
-
Plasmid DNA launches live-attenuated Japanese encephalitis virus and elicits virus-neutralizing antibodies in BALB/c mice.Virology. 2017 Dec;512:66-73. doi: 10.1016/j.virol.2017.09.005. Epub 2017 Sep 19. Virology. 2017. PMID: 28938099 Free PMC article.
-
Recent emergence of cephalosporin-resistant Salmonella Typhi in India due to the endemic clone acquiring IncFIB(K) plasmid encoding blaCTX-M-15 gene.Microbiol Spectr. 2025 Apr 10;13(5):e0087524. doi: 10.1128/spectrum.00875-24. Online ahead of print. Microbiol Spectr. 2025. PMID: 40208005 Free PMC article.
-
Isolation and sequence analysis of pCS36-4CPA, a small plasmid from Citrobacter sp. 36-4CPA.Saudi J Biol Sci. 2018 May;25(4):660-671. doi: 10.1016/j.sjbs.2016.02.014. Epub 2016 Feb 12. Saudi J Biol Sci. 2018. PMID: 29736141 Free PMC article.
-
Mechanisms of Theta Plasmid Replication.Microbiol Spectr. 2015 Feb;3(1):PLAS-0029-2014. Microbiol Spectr. 2015. PMID: 26005599 Free PMC article.
References
-
- Avgustin, J. A., and M. Grabnar. 2007. Sequence analysis of the plasmid pColG from the Escherichia coli strain CA46. Plasmid 57:89-93. - PubMed
-
- Barany, F. 1985. Single-stranded hexameric linkers: a system for in-phase insertion mutagenesis and protein engineering. Gene 37:111-123. - PubMed
-
- Benoff, B., H. Yang, C. L. Lawson, G. Parkinson, J. Liu, E. Blatter, Y. W. Ebright, H. M. Berman, and R. H. Ebright. 2002. Structural basis of transcription activation: the CAP-α CTD-DNA complex. Science 297:1562-1566. - PubMed
-
- Blaby, I. K., and D. K. Summers. 2009. The role of FIS in the Rcd checkpoint and stable maintenance of plasmid ColE1. Microbiology 155:2676-2682. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

