Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy
- PMID: 20495340
- PMCID: PMC4423623
- DOI: 10.4161/auto.6.5.12189
Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy
Abstract
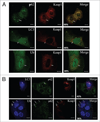
The accumulation of ubiquitin-positive protein aggregates has been implicated in the pathogenesis of neurodegenerative diseases, heart disease and diabetes. Emerging evidence indicates that the autophagy lysosomal pathway plays a critical role in the clearance of ubiquitin aggregates, a process that is mediated by the ubiquitin binding protein p62. In addition to binding ubiquitin, p62 also interacts with LC3 and transports ubiquitin conjugates to autophagosomes for degradation. The exact regulatory mechanism of this process is still largely unknown. Here we report the identification of Keap1 as a binding partner for p62 and LC3. Keap1 inhibits Nrf2 by sequestering it in the cytosol and preventing its translocation to the nucleus and activation of genes involved in the oxidative stress response. In this study, we found that Keap1 interacts with p62 and LC3 in a stress-inducible manner, and that Keap1 colocalizes with LC3 and p62 in puromycin-induced ubiquitin aggregates. Moreover, p62 serves as a bridge between Keap1 and ubiquitin aggregates and autophagosomes. Finally, genetic ablation of Keap1 leads to the accumulation of ubiquitin aggregates, increased cytotoxicity of misfolded protein aggregates, and defective activation of autophagy. Therefore, this study assigns a novel positive role of Keap1 in upregulating p62-mediated autophagic clearance of ubiquitin aggregates.
Figures





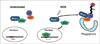
Similar articles
-
Keap1 degradation by autophagy for the maintenance of redox homeostasis.Proc Natl Acad Sci U S A. 2012 Aug 21;109(34):13561-6. doi: 10.1073/pnas.1121572109. Epub 2012 Aug 7. Proc Natl Acad Sci U S A. 2012. PMID: 22872865 Free PMC article.
-
Tripartite motif 25 inhibits protein aggregate degradation during PRRSV infection by suppressing p62-mediated autophagy.J Virol. 2024 Nov 19;98(11):e0143724. doi: 10.1128/jvi.01437-24. Epub 2024 Oct 31. J Virol. 2024. PMID: 39480084 Free PMC article.
-
Toll-Like Receptor Signaling Induces Nrf2 Pathway Activation through p62-Triggered Keap1 Degradation.Mol Cell Biol. 2015 Aug;35(15):2673-83. doi: 10.1128/MCB.00105-15. Epub 2015 May 26. Mol Cell Biol. 2015. PMID: 26012548 Free PMC article.
-
Novel target for treating Alzheimer's Diseases: Crosstalk between the Nrf2 pathway and autophagy.Ageing Res Rev. 2021 Jan;65:101207. doi: 10.1016/j.arr.2020.101207. Epub 2020 Nov 1. Ageing Res Rev. 2021. PMID: 33144123 Review.
-
p62/SQSTM1 functions as a signaling hub and an autophagy adaptor.FEBS J. 2015 Dec;282(24):4672-8. doi: 10.1111/febs.13540. Epub 2015 Oct 16. FEBS J. 2015. PMID: 26432171 Review.
Cited by
-
Depletion of the ubiquitin-binding adaptor molecule SQSTM1/p62 from macrophages harboring cftr ΔF508 mutation improves the delivery of Burkholderia cenocepacia to the autophagic machinery.J Biol Chem. 2013 Jan 18;288(3):2049-58. doi: 10.1074/jbc.M112.411728. Epub 2012 Nov 12. J Biol Chem. 2013. PMID: 23148214 Free PMC article.
-
Activation of the Keap1/Nrf2 stress response pathway in autophagic vacuolar myopathies.Acta Neuropathol Commun. 2016 Oct 31;4(1):115. doi: 10.1186/s40478-016-0384-6. Acta Neuropathol Commun. 2016. PMID: 27799074 Free PMC article.
-
Nuclear poly-glutamine aggregates rupture the nuclear envelope and hinder its repair.J Cell Biol. 2024 Nov 4;223(11):e202307142. doi: 10.1083/jcb.202307142. Epub 2024 Aug 16. J Cell Biol. 2024. PMID: 39150509 Free PMC article.
-
MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism.Breast Cancer Res Treat. 2011 Oct;129(3):983-91. doi: 10.1007/s10549-011-1604-1. Epub 2011 Jun 3. Breast Cancer Res Treat. 2011. PMID: 21638050 Free PMC article.
-
Interaction between small GTPase Rab7 and PI3KC3 links autophagy and endocytosis: A new Rab7 effector protein sheds light on membrane trafficking pathways.Small GTPases. 2011 Mar;2(2):85-88. doi: 10.4161/sgtp.2.2.15256. Small GTPases. 2011. PMID: 21776407 Free PMC article.
References
-
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. - PubMed
-
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
