Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved
- PMID: 20495566
- PMCID: PMC2883744
- DOI: 10.1038/ng.590
Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved
Abstract
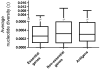
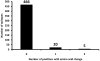
Mycobacterium tuberculosis is an obligate human pathogen capable of persisting in individual hosts for decades. We sequenced the genomes of 21 strains representative of the global diversity and six major lineages of the M. tuberculosis complex (MTBC) at 40- to 90-fold coverage using Illumina next-generation DNA sequencing. We constructed a genome-wide phylogeny based on these genome sequences. Comparative analyses of the sequences showed, as expected, that essential genes in MTBC were more evolutionarily conserved than nonessential genes. Notably, however, most of the 491 experimentally confirmed human T cell epitopes showed little sequence variation and had a lower ratio of nonsynonymous to synonymous changes than seen in essential and nonessential genes. We confirmed these findings in an additional data set consisting of 16 antigens in 99 MTBC strains. These findings are consistent with strong purifying selection acting on these epitopes, implying that MTBC might benefit from recognition by human T cells.
Conflict of interest statement
Figures

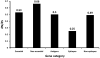
Comment in
-
Relics of selection in the mycobacterial genome.Nat Genet. 2010 Jun;42(6):476-8. doi: 10.1038/ng0610-476. Nat Genet. 2010. PMID: 20502489 Free PMC article.
References
-
- WHO. Global tuberculosis control - surveillance, planning, financing. WHO; Geneva, Switzerland: 2009.
-
- Filliol I, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006;188:759–72. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI046097/AI/NIAID NIH HHS/United States
- R01 AI051242/AI/NIAID NIH HHS/United States
- U117588500(88500)/MRC_/Medical Research Council/United Kingdom
- MC_U117588500/MRC_/Medical Research Council/United Kingdom
- R01 AI034238/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- AI046097/AI/NIAID NIH HHS/United States
- R01 AI090928/AI/NIAID NIH HHS/United States
- HHSN266200400001C/AO/NIAID NIH HHS/United States
- AI051242/AI/NIAID NIH HHS/United States
- HHSN266200700022C/PHS HHS/United States
- AI034238/AI/NIAID NIH HHS/United States

