Genetic variation in microRNA networks: the implications for cancer research
- PMID: 20495573
- PMCID: PMC2950312
- DOI: 10.1038/nrc2867
Genetic variation in microRNA networks: the implications for cancer research
Erratum in
- Nat Rev Cancer. 2010 Jul;10(7):523
Abstract
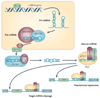
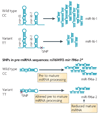
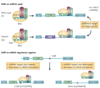
Many studies have highlighted the role that microRNAs have in physiological processes and how their deregulation can lead to cancer. More recently, it has been proposed that the presence of single nucleotide polymorphisms in microRNA genes, their processing machinery and target binding sites affects cancer risk, treatment efficacy and patient prognosis. In reviewing this new field of cancer biology, we describe the methodological approaches of these studies and make recommendations for which strategies will be most informative in the future.
Figures

Similar articles
-
[Association of human microRNA related genetic variations with cancer].Yi Chuan. 2011 Aug;33(8):870-8. doi: 10.3724/sp.j.1005.2011.00870. Yi Chuan. 2011. PMID: 21831803 Review. Chinese.
-
MicroRNAs in cancer: biomarkers, functions and therapy.Trends Mol Med. 2014 Aug;20(8):460-9. doi: 10.1016/j.molmed.2014.06.005. Epub 2014 Jul 12. Trends Mol Med. 2014. PMID: 25027972 Review.
-
SNPing cancer in the bud: microRNA and microRNA-target site polymorphisms as diagnostic and prognostic biomarkers in cancer.Pharmacol Ther. 2013 Jan;137(1):55-63. doi: 10.1016/j.pharmthera.2012.08.016. Epub 2012 Sep 3. Pharmacol Ther. 2013. PMID: 22964086 Free PMC article. Review.
-
Identification of diagnostic and prognostic biomarkers for cancer: Focusing on genetic variations in microRNA regulatory pathways (Review).Mol Med Rep. 2016 Mar;13(3):1943-52. doi: 10.3892/mmr.2016.4782. Epub 2016 Jan 14. Mol Med Rep. 2016. PMID: 26782081 Review.
-
Functional microRNA binding site variants.Mol Oncol. 2019 Jan;13(1):4-8. doi: 10.1002/1878-0261.12421. Epub 2018 Dec 26. Mol Oncol. 2019. PMID: 30536617 Free PMC article. Review.
Cited by
-
Genetic polymorphisms of miR-146a and miR-27a, H. pylori infection, and risk of gastric lesions in a Chinese population.PLoS One. 2013 Apr 17;8(4):e61250. doi: 10.1371/journal.pone.0061250. Print 2013. PLoS One. 2013. PMID: 23613822 Free PMC article.
-
miRNAs as biomarkers in prostate cancer.Clin Transl Oncol. 2012 Nov;14(11):803-11. doi: 10.1007/s12094-012-0877-0. Epub 2012 Jul 24. Clin Transl Oncol. 2012. PMID: 22855165 Review.
-
rs4919510 in hsa-mir-608 is associated with outcome but not risk of colorectal cancer.PLoS One. 2012;7(5):e36306. doi: 10.1371/journal.pone.0036306. Epub 2012 May 11. PLoS One. 2012. PMID: 22606253 Free PMC article.
-
Single nucleotide polymorphisms in microRNA binding sites: implications in colorectal cancer.ScientificWorldJournal. 2014;2014:547154. doi: 10.1155/2014/547154. Epub 2014 Dec 24. ScientificWorldJournal. 2014. PMID: 25654126 Free PMC article.
-
hsa-miR-423 rs6505162 Is Associated with The Increased Risk of Breast Cancer in Isfahan Central Province of Iran.Cell J. 2020 Jul;22(Suppl 1):110-116. doi: 10.22074/cellj.2020.7011. Epub 2020 Jul 18. Cell J. 2020. PMID: 32779440 Free PMC article.
References
-
-
Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861.This paper provides a good background to HapMap and provides details of the most recent version of the database.
-
-
-
Kan YW, Dozy AM. Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proc. Natl Acad. Sci. USA. 1978;75:5631–5635.A seminal study that describes the first detection of SNPs
-
-
-
Bartel B. MicroRNAs directing siRNA biogenesis. Nature Struct. Mol. Biol. 2005;12:569–571.This is an excellent paper that covers miRNA biogenesis in a complete and descriptive manner.
-
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

